PythonCall & JuliaCall
Bringing Python® and Julia together in seamless harmony:
- Call Python code from Julia and Julia code from Python via a symmetric interface.
- Simple syntax, so the Python code looks like Python and the Julia code looks like Julia.
- Intuitive and flexible conversions between Julia and Python: anything can be converted, you are in control.
- Fast non-copying conversion of numeric arrays in either direction: modify Python arrays (e.g.
bytes,array.array,numpy.ndarray) from Julia or Julia arrays from Python. - Helpful wrappers: interpret Python sequences, dictionaries, arrays, dataframes and IO streams as their Julia couterparts, and vice versa.
- Beautiful stack-traces.
- Supports modern systems: tested on Windows, MacOS and Linux, 64-bit, Julia 1.4 upwards and Python 3.5 upwards.
To get started, read the documentation.
Example 1: Calling Python from Julia
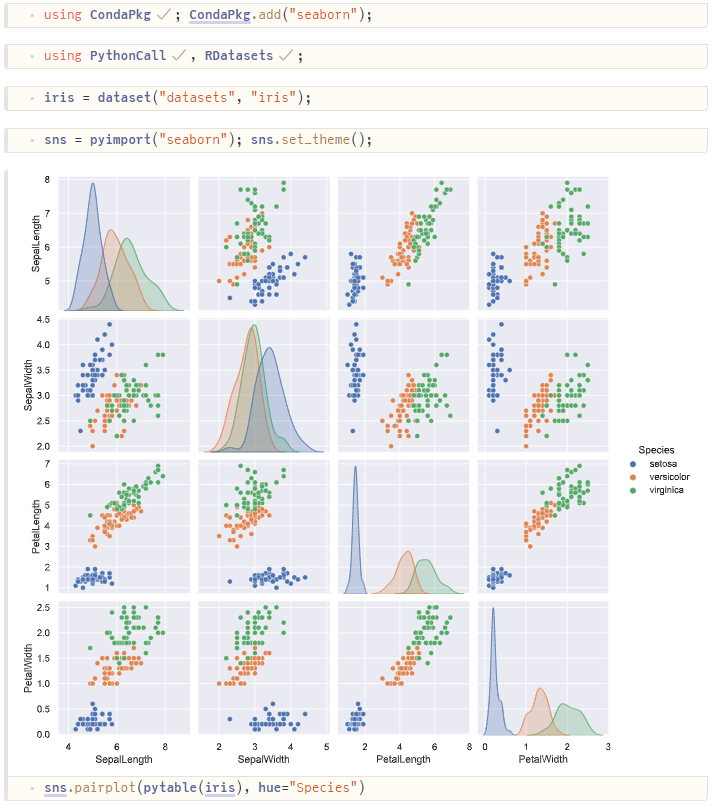
In this example, we use the Julia module PythonCall from a Pluto notebook to inspect the Iris dataset:
- We load the Iris dataset as a Julia DataFrame using RDatasets.
- We use
pytable(df)to convert it to a Python Pandas DataFrame. - We use the Python package Seaborn to produce a pair-plot, which is automatically displayed.
Example 2: Calling Julia from Python
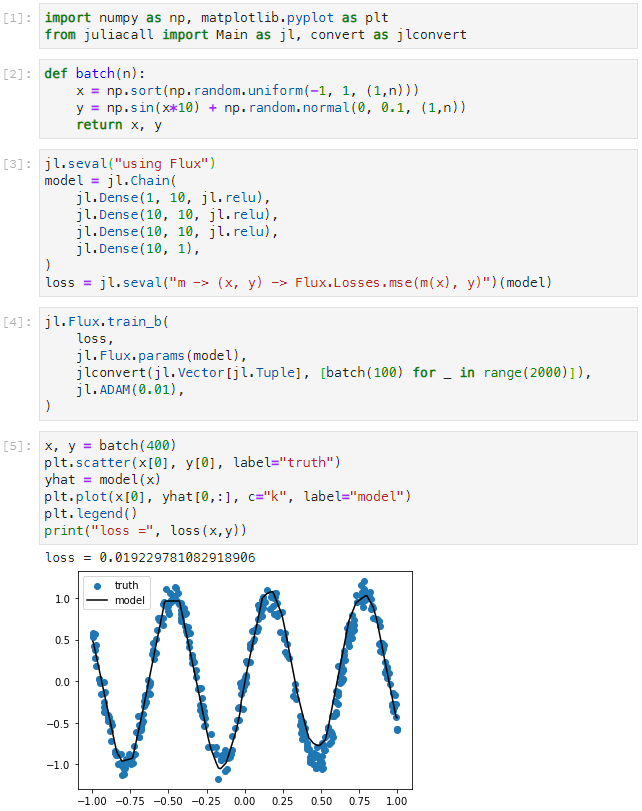
In this example we use the Python module JuliaCall from an IPython notebook to train a simple neural network:
- We generate some random training data using Python's Numpy.
- We construct and train a neural network model using Julia's Flux.
- We plot some sample output from the model using Python's MatPlotLib.
What about PyCall?
The existing package PyCall is another similar interface to Python. Here we note some key differences, but a more detailed comparison is in the documentation.
- PythonCall supports a wider range of conversions between Julia and Python, and the conversion mechanism is extensible.
- PythonCall by default never copies mutable objects when converting, but instead directly wraps the mutable object. This means that modifying the converted object modifies the original, and conversion is faster.
- PythonCall does not usually automatically convert results to Julia values, but leaves them as Python objects. This makes it easier to do Pythonic things with these objects (e.g. accessing methods) and is type-stable.
- PythonCall installs dependencies into a separate conda environment for each Julia project. This means each Julia project can have an isolated set of Python dependencies.
- PythonCall supports Julia 1.0+ and Python 3.5+ whereas PyCall supports Julia 0.7+ and Python 2.7+.