This PR convert the existing test modules to use pytest for test discovery and running. Tests were also updated for minor cleanup and to utilize pytest features (e.g. parameterize and fixtures) effectively. The end result is to afford testing options that are more familiar and ergonomic to "standard" python devs.
Overview
Cleanup
A few commits perform some minor cleanup on the existing tests:
- https://github.com/nv-legate/cunumeric/pull/297/commits/572e7f106cffb80df2d0dcd0bb17ff423d2e2b11 — remove old and outdated "universal functions" tests
- https://github.com/nv-legate/cunumeric/pull/297/commits/5e0ff856667234f0bff4d0ff48ab08dcf192ed7c — remove a useless test
- https://github.com/nv-legate/cunumeric/pull/297/commits/5d8a6172caa97501e795688c22d969efbe404056 — remove redundant bare
return statements throughout
File movement
-
To make it easier to use pytest for test selection, the existing tests were moved and renamed:
-
https://github.com/nv-legate/cunumeric/pull/297/commits/3f3c5cb8d3ed05a0981990e9920d851d58dd19a1 — move files to integration subdirectory
-
https://github.com/nv-legate/cunumeric/pull/297/commits/9f4f4108449f56f360e90c2f56d30569796b7c0c — prefix all test module filenames with test_
Moving the files to a subdirectory allows this entire group of tests to be easily run by executing pytest with this directory path. The test_ prefix is to conform with pytest expectations for default test discovery.
Pytest updates
- https://github.com/nv-legate/cunumeric/pull/297/commits/571ea075d549da3875550aa0d28f4154af83339d — rename some helper functions to avoid
test_ prefix. By default, pytest will interpret anything starting with test or Test as test to run.
- https://github.com/nv-legate/cunumeric/pull/297/commits/76779365e25cff1c9274b17f454145267aee51b8 — a minimal update that adds
pytest scaffolding to each test module and the least change to get running. For most tests this was just a couple of lines of boilerplate change. But a few tests required slightly more extensive updates.
- https://github.com/nv-legate/cunumeric/pull/297/commits/bbda5a96336d320ade26e34cd6359acc6da6d0b7 — more in-depth changed to split out tests into sensible smaller jobs, and to utilize parameterization and fixtures.
Notes
Further notes will be inline.
Operation
Running test.py
The existing way to run tests with test.py is unchanged:
./test.py --use=cuda
This still generates the existing report:
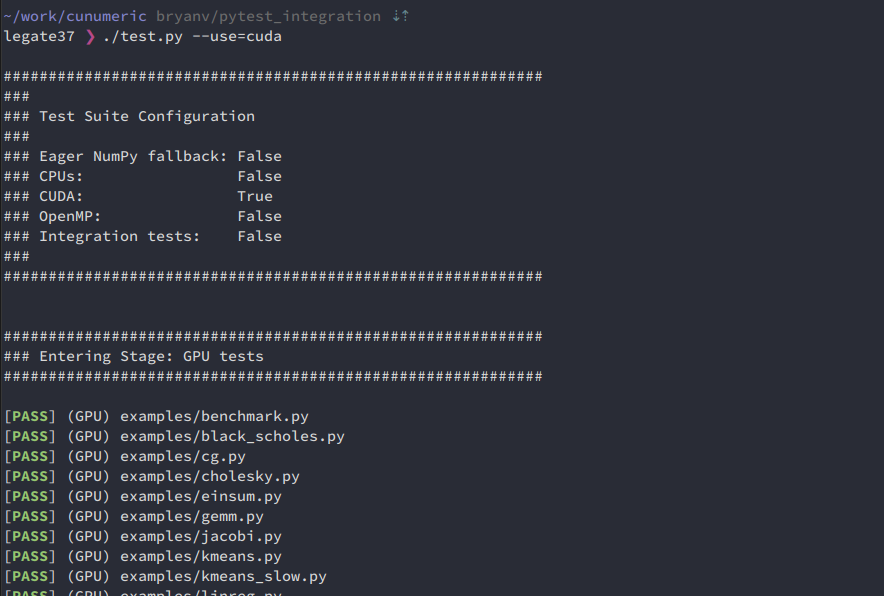
~~(Note: Currently test.py -v only shows stdout from test.py and not from tests. This will be straightforward to restore but I would like to do that in a follow-on PR dedicated to test.py. Test stdout can be observed with either of the methods below with the standard -s flag. cc @magnatelee)~~
Executing individual tests
Individual tests can still be executed by running the module with legate including by passing in runtime command line options:
legate tests/integration/test_nonzero.py --gpus 2 -cunumeric:test
However, now the output is a standard pytest report:
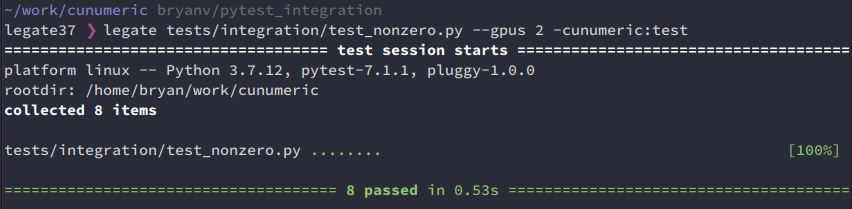
It is also now possible to pass standard pytests options, e.g. -v for a verbose report:
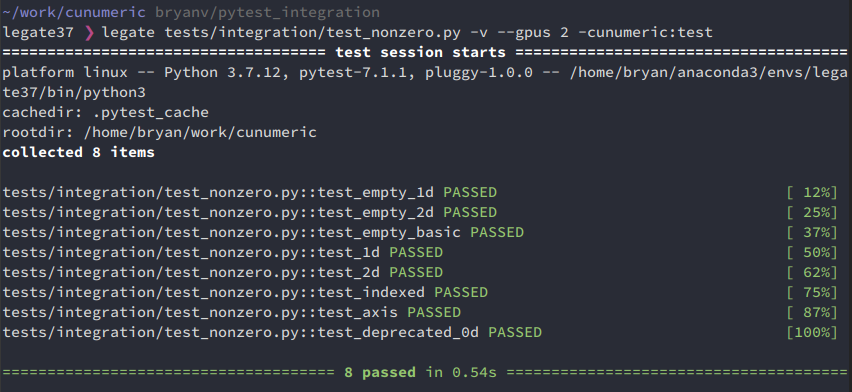
Using pytest for test discovery
Although it is not yet possible to simply run pytest <dir> in the typical way, it is possible to achieve the same operation with a little more explicit invocation:
legate -c "import pytest; pytest.main(['tests/integration'])" --gpus 2 -cunumeric:test
Note that all the standard pytest options, e.g. incluing -k and -m for test filtering, can be used.
The above will run all the tests under tests/integration, and generate a standard combined pytest report:
The full report output text can be seen here:
legate37 ❯ legate -c "import pytest; pytest.main(['tests/integration'])" --gpus 2 -cunumeric:test
WARNING: Disabling control replication for interactive run
========================================================================================= test session starts ==========================================================================================
platform linux -- Python 3.7.12, pytest-7.1.1, pluggy-1.0.0
rootdir: /home/bryan/work/cunumeric
collected 2860 items
tests/integration/test_2d_reduction.py ... [ 0%]
tests/integration/test_3d_reduction.py . [ 0%]
tests/integration/test_advanced_indexing.py . [ 0%]
tests/integration/test_append.py ........ [ 0%]
tests/integration/test_argmin.py .. [ 0%]
tests/integration/test_array_creation.py .............. [ 1%]
tests/integration/test_array_split.py ....... [ 1%]
tests/integration/test_binary_op.py ..... [ 1%]
tests/integration/test_binary_op_2d.py .... [ 1%]
tests/integration/test_binary_op_broadcast.py .... [ 1%]
tests/integration/test_binary_op_complex.py ..... [ 1%]
tests/integration/test_binary_op_typing.py ..................................................................................................................................................... [ 7%]
................................................................................................................................................................................................ [ 13%]
................................................................................................................................................................................................ [ 20%]
................................................................................................................................................................................................ [ 27%]
................................................................................................................................................................................................ [ 33%]
................................................................................................................................................................................................ [ 40%]
................................................................................................................................................................................................ [ 47%]
................................................................................................................................................................................................ [ 54%]
................................................................................................................................................................................................ [ 60%]
................................................................................................................................................................................................ [ 67%]
................................................................................................................................................................................................ [ 74%]
................................................................................................................................................................................. [ 80%]
tests/integration/test_binary_ufunc.py . [ 80%]
tests/integration/test_bincount.py ...... [ 80%]
tests/integration/test_block.py ............ [ 81%]
tests/integration/test_cholesky.py ......... [ 81%]
tests/integration/test_compare.py ...... [ 81%]
tests/integration/test_complex_ops.py .... [ 81%]
tests/integration/test_concatenate_stack.py ................................................ [ 83%]
tests/integration/test_contains.py .. [ 83%]
tests/integration/test_convolve.py ...... [ 83%]
tests/integration/test_copy.py . [ 83%]
tests/integration/test_dot.py ......................... [ 84%]
tests/integration/test_einsum.py ....................................................................................................................................................... [ 89%]
tests/integration/test_eye.py ............... [ 90%]
tests/integration/test_fill.py . [ 90%]
tests/integration/test_flatten.py ......... [ 90%]
tests/integration/test_flip.py .......... [ 91%]
tests/integration/test_get_item.py . [ 91%]
tests/integration/test_index_routines.py ............ [ 91%]
tests/integration/test_ingest.py .... [ 91%]
tests/integration/test_inlinemap-keeps-region-alive.py . [ 91%]
tests/integration/test_inner.py ......................... [ 92%]
tests/integration/test_interop.py . [ 92%]
tests/integration/test_intra_array_copy.py .... [ 92%]
tests/integration/test_jacobi.py . [ 92%]
tests/integration/test_length.py .... [ 92%]
tests/integration/test_linspace.py .... [ 93%]
tests/integration/test_logical.py .....s [ 93%]
tests/integration/test_lstm_backward_test.py . [ 93%]
tests/integration/test_lstm_simple_forward.py . [ 93%]
tests/integration/test_map_reduce.py . [ 93%]
tests/integration/test_mask.py .... [ 93%]
tests/integration/test_matmul.py ................ [ 94%]
tests/integration/test_nonzero.py ........ [ 94%]
tests/integration/test_norm.py . [ 94%]
tests/integration/test_numpy_interop.py . [ 94%]
tests/integration/test_outer.py ................. [ 95%]
tests/integration/test_overwrite_slice.py . [ 95%]
tests/integration/test_randint.py .. [ 95%]
tests/integration/test_reduction.py ....... [ 95%]
tests/integration/test_reduction_axis.py .................. [ 96%]
tests/integration/test_reduction_complex.py .. [ 96%]
tests/integration/test_repeat.py ...... [ 96%]
tests/integration/test_reshape.py ................. [ 96%]
tests/integration/test_set_item.py . [ 96%]
tests/integration/test_shape.py .s [ 97%]
tests/integration/test_singleton_access.py . [ 97%]
tests/integration/test_slicing.py ..ss [ 97%]
tests/integration/test_sort.py . [ 97%]
tests/integration/test_squeeze.py . [ 97%]
tests/integration/test_swapaxes.py .... [ 97%]
tests/integration/test_tensordot.py ......................... [ 98%]
tests/integration/test_tile.py .. [ 98%]
tests/integration/test_transpose.py .. [ 98%]
tests/integration/test_trilu.py .......... [ 98%]
tests/integration/test_unary_functions_2d.py ........ [ 99%]
tests/integration/test_unary_functions_2d_complex.py ..... [ 99%]
tests/integration/test_unary_ufunc.py . [ 99%]
tests/integration/test_unique.py ..... [ 99%]
tests/integration/test_update.py . [ 99%]
tests/integration/test_vdot.py .... [ 99%]
tests/integration/test_view.py . [ 99%]
tests/integration/test_vstack.py ... [ 99%]
tests/integration/test_where.py .....s [ 99%]
tests/integration/test_window.py . [100%]
=========================================================================================== warnings summary ===========================================================================================
tests/integration/test_advanced_indexing.py: 124 warnings
/home/bryan/work/cunumeric/cunumeric/array.py:773: DeprecationWarning: `np.bool` is a deprecated alias for the builtin `bool`. To silence this warning, use `bool` by itself. Doing this will not modify any behavior and is safe. If you specifically wanted the numpy scalar type, use `np.bool_` here.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
if key.dtype != np.bool and not np.issubdtype(
tests/integration/test_advanced_indexing.py: 124 warnings
/home/bryan/work/cunumeric/cunumeric/array.py:777: DeprecationWarning: `np.bool` is a deprecated alias for the builtin `bool`. To silence this warning, use `bool` by itself. Doing this will not modify any behavior and is safe. If you specifically wanted the numpy scalar type, use `np.bool_` here.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
if key.dtype != np.bool and key.dtype != np.int64:
tests/integration/test_advanced_indexing.py: 30 warnings
/home/bryan/work/cunumeric/cunumeric/deferred.py:425: DeprecationWarning: `np.bool` is a deprecated alias for the builtin `bool`. To silence this warning, use `bool` by itself. Doing this will not modify any behavior and is safe. If you specifically wanted the numpy scalar type, use `np.bool_` here.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
and key.dtype == np.bool
tests/integration/test_advanced_indexing.py: 120 warnings
/home/bryan/work/cunumeric/cunumeric/deferred.py:479: DeprecationWarning: `np.bool` is a deprecated alias for the builtin `bool`. To silence this warning, use `bool` by itself. Doing this will not modify any behavior and is safe. If you specifically wanted the numpy scalar type, use `np.bool_` here.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
and k.dtype == np.bool
tests/integration/test_advanced_indexing.py: 120 warnings
/home/bryan/work/cunumeric/cunumeric/deferred.py:516: DeprecationWarning: `np.bool` is a deprecated alias for the builtin `bool`. To silence this warning, use `bool` by itself. Doing this will not modify any behavior and is safe. If you specifically wanted the numpy scalar type, use `np.bool_` here.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
if k.dtype == np.bool:
tests/integration/test_advanced_indexing.py::test
/home/bryan/work/cunumeric/tests/integration/test_advanced_indexing.py:118: RuntimeWarning: converting index array to int64 type
assert np.array_equal(x[index], x_num[index_num])
tests/integration/test_advanced_indexing.py::test
/home/bryan/work/cunumeric/tests/integration/test_advanced_indexing.py:122: RuntimeWarning: converting index array to int64 type
x_num[index_num] = 3.5
tests/integration/test_advanced_indexing.py::test
/home/bryan/work/cunumeric/tests/integration/test_advanced_indexing.py:129: UserWarning: cuNumeric performing implicit type conversion from float64 to int64
x_num[index_num] = b_num
tests/integration/test_advanced_indexing.py::test
/home/bryan/work/cunumeric/tests/integration/test_advanced_indexing.py:129: RuntimeWarning: converting index array to int64 type
x_num[index_num] = b_num
tests/integration/test_advanced_indexing.py::test
/home/bryan/work/cunumeric/tests/integration/test_advanced_indexing.py:559: UserWarning: cuNumeric performing implicit type conversion from float64 to int64
z_num[indx_num] = b_num
tests/integration/test_advanced_indexing.py::test
/home/bryan/work/cunumeric/tests/integration/test_advanced_indexing.py:609: RuntimeWarning: converting index array to int64 type
res_num = x_num[ind_num, ind_num]
tests/integration/test_advanced_indexing.py::test
/home/bryan/work/cunumeric/tests/integration/test_advanced_indexing.py:627: UserWarning: cuNumeric performing implicit type conversion from int16 to float64
x_num[ind_num, ind_num] = b_num
tests/integration/test_cholesky.py::test_complex[8]
tests/integration/test_cholesky.py::test_complex[9]
tests/integration/test_cholesky.py::test_complex[255]
tests/integration/test_cholesky.py::test_complex[512]
/home/bryan/work/cunumeric/tests/integration/test_cholesky.py:48: UserWarning: cuNumeric performing implicit type conversion from complex128 to float64
d[1] = b
tests/integration/test_dot.py::test_dot[1-1]
tests/integration/test_dot.py::test_dot[1-2]
tests/integration/test_dot.py::test_dot[2-1]
tests/integration/test_dot.py::test_dot[2-2]
/home/bryan/work/cunumeric/tests/integration/test_dot.py:34: UserWarning: cuNumeric performing implicit type conversion from float16 to float32
return lib.dot(*args, **kwargs)
tests/integration/test_einsum.py::test_cast[->]
tests/integration/test_einsum.py::test_cast[a->]
tests/integration/test_einsum.py::test_cast[a,->]
tests/integration/test_einsum.py::test_cast[a,a->]
/home/bryan/work/cunumeric/cunumeric/array.py:123: ComplexWarning: Casting complex values to real discards the imaginary part
*args, **kwargs
tests/integration/test_einsum.py::test_cast[a->a]
tests/integration/test_einsum.py::test_cast[a,->a]
tests/integration/test_einsum.py::test_cast[a,a->]
tests/integration/test_einsum.py::test_cast[a,a->a]
tests/integration/test_einsum.py::test_cast[a,b->ab]
tests/integration/test_einsum.py::test_cast[ab,ca->a]
tests/integration/test_einsum.py::test_cast[ab,ca->b]
/home/bryan/work/cunumeric/tests/integration/test_einsum.py:235: UserWarning: cuNumeric performing implicit type conversion from float16 to float32
cn.einsum(expr, *cn_inputs, out=cn_out)
tests/integration/test_einsum.py::test_cast[a->a]
tests/integration/test_einsum.py::test_cast[a,->a]
tests/integration/test_einsum.py::test_cast[a,a->]
tests/integration/test_einsum.py::test_cast[a,a->a]
tests/integration/test_einsum.py::test_cast[a,b->ab]
tests/integration/test_einsum.py::test_cast[ab,ca->a]
tests/integration/test_einsum.py::test_cast[ab,ca->b]
/home/bryan/work/cunumeric/tests/integration/test_einsum.py:235: UserWarning: cuNumeric performing implicit type conversion from float16 to complex64
cn.einsum(expr, *cn_inputs, out=cn_out)
tests/integration/test_einsum.py::test_cast[a->a]
tests/integration/test_einsum.py::test_cast[a,->a]
tests/integration/test_einsum.py::test_cast[a,a->a]
tests/integration/test_einsum.py::test_cast[a,b->ab]
tests/integration/test_einsum.py::test_cast[ab,ca->a]
tests/integration/test_einsum.py::test_cast[ab,ca->b]
/home/bryan/work/cunumeric/tests/integration/test_einsum.py:235: UserWarning: cuNumeric performing implicit type conversion from float32 to float16
cn.einsum(expr, *cn_inputs, out=cn_out)
tests/integration/test_einsum.py::test_cast[a->a]
tests/integration/test_einsum.py::test_cast[a,->a]
tests/integration/test_einsum.py::test_cast[a,a->]
tests/integration/test_einsum.py::test_cast[a,a->a]
tests/integration/test_einsum.py::test_cast[a,b->ab]
tests/integration/test_einsum.py::test_cast[ab,ca->a]
tests/integration/test_einsum.py::test_cast[ab,ca->b]
/home/bryan/work/cunumeric/tests/integration/test_einsum.py:235: UserWarning: cuNumeric performing implicit type conversion from float32 to complex64
cn.einsum(expr, *cn_inputs, out=cn_out)
tests/integration/test_einsum.py::test_cast[a->a]
tests/integration/test_einsum.py::test_cast[a,->a]
tests/integration/test_einsum.py::test_cast[a,a->a]
tests/integration/test_einsum.py::test_cast[a,b->ab]
tests/integration/test_einsum.py::test_cast[ab,ca->a]
tests/integration/test_einsum.py::test_cast[ab,ca->b]
/home/bryan/work/cunumeric/tests/integration/test_einsum.py:235: UserWarning: cuNumeric performing implicit type conversion from complex64 to float16
cn.einsum(expr, *cn_inputs, out=cn_out)
tests/integration/test_einsum.py::test_cast[a->a]
tests/integration/test_einsum.py::test_cast[a,->a]
tests/integration/test_einsum.py::test_cast[a,a->a]
tests/integration/test_einsum.py::test_cast[a,b->ab]
tests/integration/test_einsum.py::test_cast[ab,ca->a]
tests/integration/test_einsum.py::test_cast[ab,ca->b]
/home/bryan/work/cunumeric/tests/integration/test_einsum.py:235: UserWarning: cuNumeric performing implicit type conversion from complex64 to float32
cn.einsum(expr, *cn_inputs, out=cn_out)
tests/integration/test_einsum.py::test_cast[a,a->]
tests/integration/test_einsum.py::test_cast[a,a->a]
tests/integration/test_einsum.py::test_cast[a,b->ab]
tests/integration/test_einsum.py::test_cast[ab,ca->a]
tests/integration/test_einsum.py::test_cast[ab,ca->b]
/home/bryan/work/cunumeric/tests/integration/test_einsum.py:228: UserWarning: cuNumeric performing implicit type conversion from float16 to float32
cn_res = cn.einsum(expr, *cn_inputs)
tests/integration/test_einsum.py::test_cast[a,a->]
tests/integration/test_einsum.py::test_cast[a,a->a]
tests/integration/test_einsum.py::test_cast[a,b->ab]
tests/integration/test_einsum.py::test_cast[ab,ca->a]
tests/integration/test_einsum.py::test_cast[ab,ca->b]
/home/bryan/work/cunumeric/tests/integration/test_einsum.py:228: UserWarning: cuNumeric performing implicit type conversion from float16 to complex64
cn_res = cn.einsum(expr, *cn_inputs)
tests/integration/test_einsum.py::test_cast[a,a->]
tests/integration/test_einsum.py::test_cast[a,a->a]
tests/integration/test_einsum.py::test_cast[a,b->ab]
tests/integration/test_einsum.py::test_cast[ab,ca->a]
tests/integration/test_einsum.py::test_cast[ab,ca->b]
/home/bryan/work/cunumeric/tests/integration/test_einsum.py:228: UserWarning: cuNumeric performing implicit type conversion from float32 to complex64
cn_res = cn.einsum(expr, *cn_inputs)
tests/integration/test_flatten.py::test_basic[(1, 1)]
tests/integration/test_flatten.py::test_basic[(1, 1, 1)]
tests/integration/test_flatten.py::test_basic[(1, 10)]
tests/integration/test_flatten.py::test_basic[(1, 10, 1)]
tests/integration/test_flatten.py::test_basic[(10, 10)]
tests/integration/test_flatten.py::test_basic[(10, 10, 10)]
/home/bryan/work/cunumeric/tests/integration/test_flatten.py:26: RuntimeWarning: cuNumeric has not implemented reshape using Fortran-like index order and is falling back to canonical numpy. You may notice significantly decreased performance for this function call.
c = num_arr.flatten(order)
tests/integration/test_index_routines.py::test_choose_1d
/home/bryan/work/cunumeric/tests/integration/test_index_routines.py:42: UserWarning: cuNumeric performing implicit type conversion from int64 to float64
assert np.array_equal(
tests/integration/test_sort.py::test
/home/bryan/work/cunumeric/tests/integration/test_sort.py:23: UserWarning: cuNumeric performing implicit type conversion from complex64 to complex128
if not num.allclose(a_np, a_num):
tests/integration/test_vdot.py::test[complex64-complex128]
tests/integration/test_vdot.py::test[complex128-complex64]
/home/bryan/work/cunumeric/tests/integration/test_vdot.py:28: UserWarning: cuNumeric performing implicit type conversion from complex64 to complex128
mk_0to1_array(lib, (5,), dtype=b_dtype),
-- Docs: https://docs.pytest.org/en/stable/how-to/capture-warnings.html
============================================================================ 2855 passed, 5 skipped, 601 warnings in 41.34s ============================================================================
There are some warning that show up in the output at the end, mostly about implicit casting. I am not sure if these are expected or not (I expect they would be)
Future work
Things that are not in this PR but are planned for follow-on PRs:
- Using
pytest to run the examples
- ~~Restoring previous
-v behaviour to test.py~~
- Increased documentation for running tests
- Allowing to run with simpler
legate -m pytest




