This is the release note of v3.0.0.
This is not something you have to read from top to bottom to learn about the summary of Optuna v3. The recommended way is reading the release blog.
If you want to update your existing projects from Optuna v2.x to Optuna v3, please see the migration guide and try out Optuna v3.
Highlights
New Features
New NSGA-II Crossover Options
New crossover options are added to NSGA-II sampler, the default multi-objective algorithm of Optuna. The performance for floating point parameters are improved. Please visit #2903, #3221, and the document for more information.

A New Algorithm: Quasi-Monte Carlo Sampler
Quasi-Monte Carlo sampler is now supported. It can be used in place of RandomSampler, and can improve performance especially for high dimensional problems. See #2423, #2964, and the document for more information.

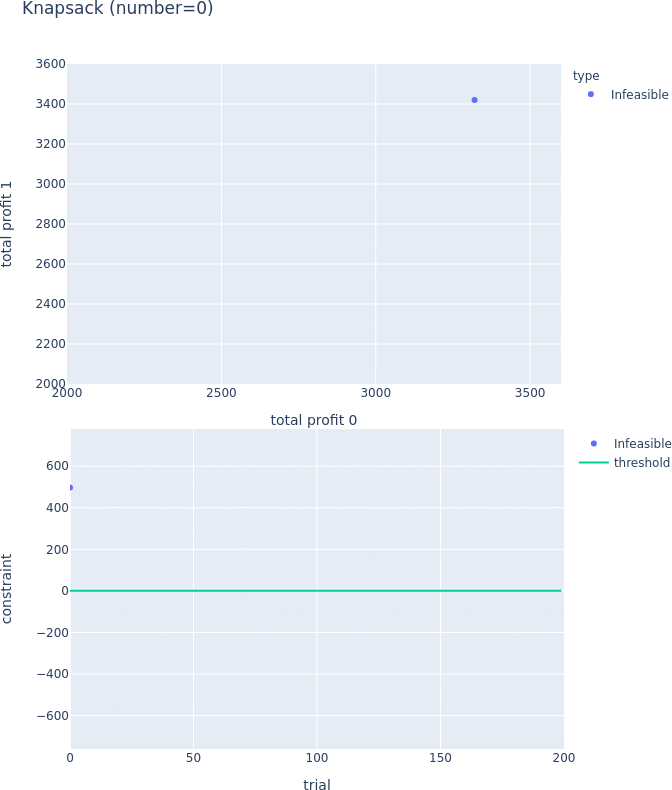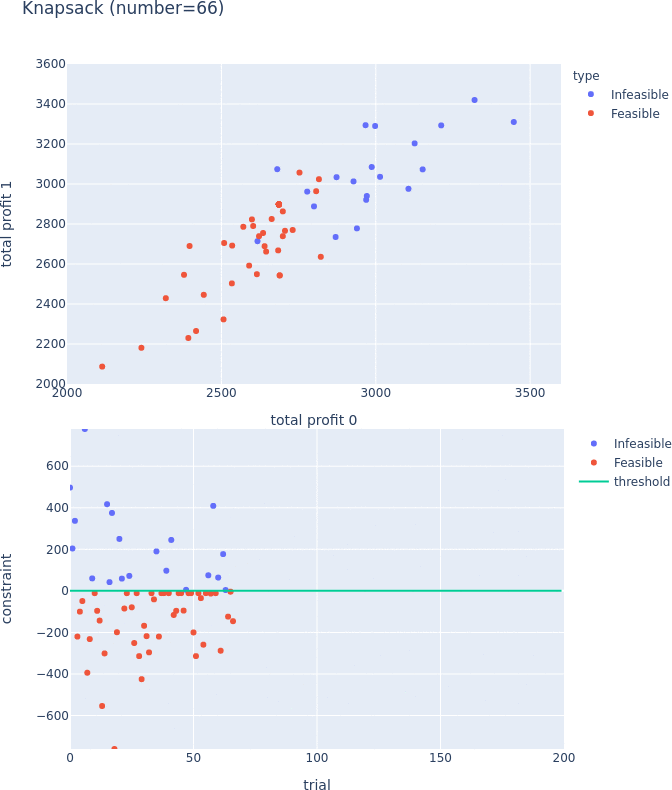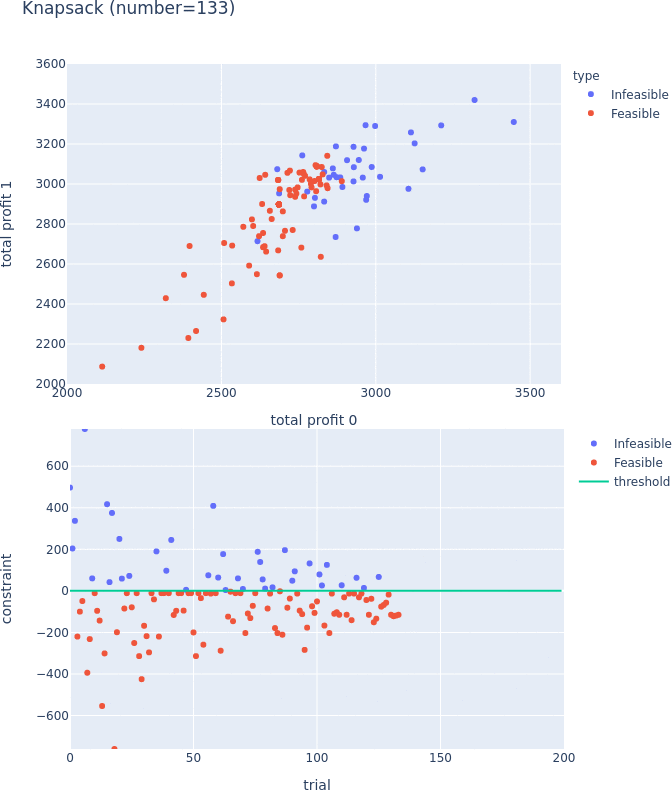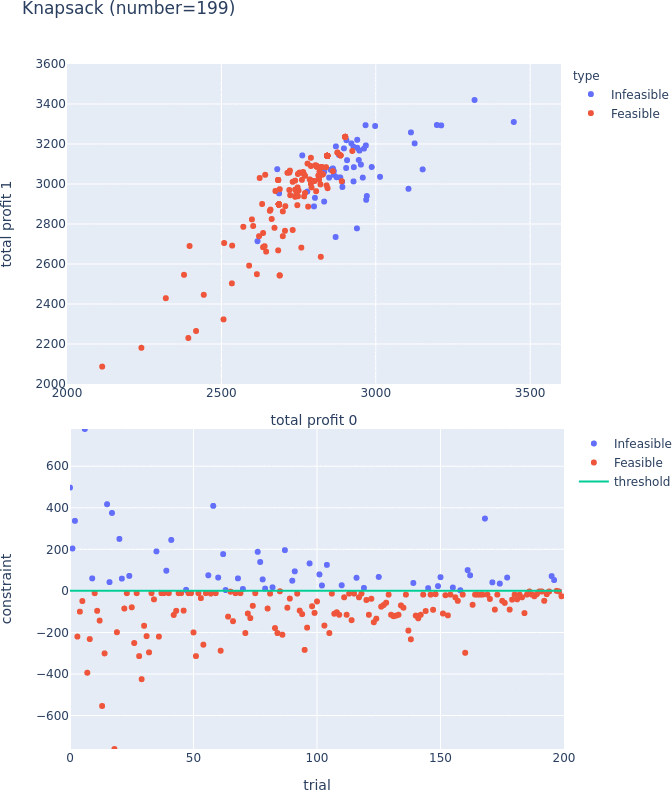
Constrained Optimization Support for TPE
TPESampler now supports constraint-aware optimization. For more information on this feature, please visit #3506 and the document.
| Without constraints | With constraints |
| - | - |
|  |
|  |
|
Constraints Support for Pareto-front Plot
Pareto-front plot now shows which trials satisfy the constraints and which do not. For more information, please see the following PRs (#3128, #3497, and #3389) and the document.

A New Importance Evaluator: ShapleyImportanceEvaluator
We introduced a new importance evaluator, optuna.integration.ShapleyImportanceEvaluator, which uses SHAP. See #3507 and the document for more information.

New History Visualization with Multiple Studies
Optimization history plot can now compare multiple studies or display the mean and variance of multiple studies optimized with the same settings. For more information, please see the following multiple PRs (#2807, #3062, #3122, and #3736) and the document.


Improved Stability
Optuna has a number of core APIs. One being the suggest API and the optuna.Study class. The visualization module is also frequently used to analyze results. Many of these have been simplified, stabilized, and refactored in v3.0.
Simplified Suggest API
The suggest API has been aggregated into 3 APIs: suggest_float for floating point parameters, suggest_int for integer parameters, and suggest_catagorical for categorical parameters. For more information, see #2939, #2941, and PRs submitted for those issues.
Introduction of a Test Policy
We have developed and published a test policy in v3.0 that defines how tests for Optuna should be written. Based on the published test policy, we have improved many unit tests. For more information, see https://github.com/optuna/optuna/issues/2974 and PRs with test label.
Visualization Refactoring
Optuna's visualization module had a deep history and various debts. We have worked throughout v3.0 to eliminate this debt with the help of many contributors. See #2893, #2913, #2959 and PRs submitted for those issues.
Stabilized Features
Through the development of v3.0, we have decided to provide many experimental features as stable features by going through their behavior, fixing bugs, and analyzing use cases. The following is a list of features that have been stabilized in v3.0.
Performance Verification
Optuna has many algorithms implemented, but many of their behaviors and characteristics are unknown to the user. We have developed the following table to inform users of empirically known behaviors and characteristics. See #3571 and #3593 for more details.

To quantitatively assess the performance of our algorithms, we have developed a benchmarking environment. We also evaluated the performance of the algorithms by conducting actual benchmarking experiments using this environment. See here, #2964, and #2906 for more details.

Breaking Changes
Changes to the RDB schema:
- To use Optuna v3.0.0 with
RDBStorage that was created in the previous versions of Optuna, please execute optuna storage upgrade to migrate your database (#3113, #3559, #3603, #3668).
Features deprecated in 3.0:
suggest_uniform(), suggest_loguniform(), and suggest_discrete_uniform()
UniformDistribution, LogUniformDistribution, DiscreteUniformDistribution, IntUniformDistribution, and IntLogUniformDistribution (#3246, #3420)- Positional arguments of
create_study(), load_study(), delete_study(), and create_study() (#3270)
axis_order argument of plot_pareto_front() (#3341)
Features removed in 3.0:
optuna dashboard command (#3058)optuna.structs module (#3057)best_booster property of LightGBMTuner (#3057)type_checking module (#3235)
Minor breaking changes:
- Add option to exclude best trials from study summaries (#3109)
- Move validation logic from
_run_trial to study.tell (#3144)
- Use an enqueued parameter that is out of range from suggest API (#3298)
- Fix distribution compatibility for linear and logarithmic distribution (#3444)
- Remove
get_study_id_from_trial_id, the method of BaseStorage (#3538)
New Features
- Add interval for LightGBM callback (#2490)
- Allow multiple studies and add error bar option to
plot_optimization_history (#2807)
- Support PyTorch-lightning DDP training (#2849, thanks @tohmae!)
- Add crossover operators for NSGA-II (#2903, thanks @yoshinobc!)
- Add abbreviated JSON formats of distributions (#2905)
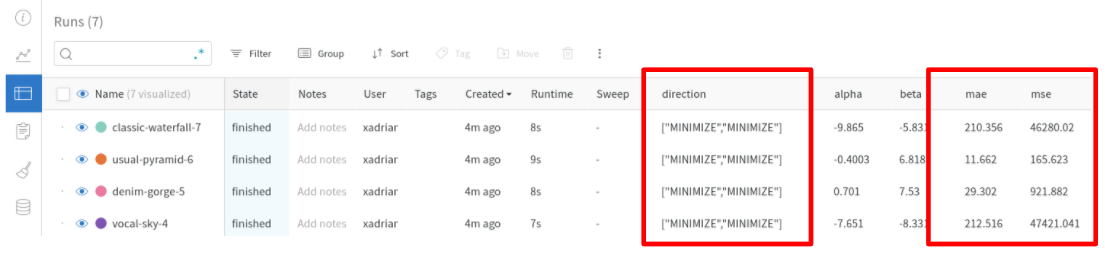
- Extend
MLflowCallback interface (#2912, thanks @xadrianzetx!)
- Support AllenNLP distributed pruning (#2977)
- Make
trial.user_attrs logging optional in MLflowCallback (#3043, thanks @xadrianzetx!)
- Support multiple input of studies when plot with Matplotlib (#3062, thanks @TakuyaInoue-github!)
- Add
IntDistribution & FloatDistribution (#3063, thanks @nyanhi!)
- Add
trial.user_attrs to pareto_front hover text (#3082, thanks @kasparthommen!)
- Support error bar for Matplotlib (#3122, thanks @TakuyaInoue-github!)
- Add
optuna tell with --skip-if-finished (#3131)
- Add QMC sampler (#2423, thanks @kstoneriv3!)
- Refactor pareto front and support
constraints_func in plot_pareto_front (#3128, thanks @semiexp!)
- Add
skip_if_finished flag to Study.tell (#3150, thanks @xadrianzetx!)
- Add
user_attrs argument to Study.enqueue_trial (#3185, thanks @knshnb!)
- Option to inherit intermediate values in
RetryFailedTrialCallback (#3269, thanks @knshnb!)
- Add setter method for
DiscreteUniformDistribution.q (#3283)
- Stabilize allennlp integrations (#3228)
- Stabilize
create_trial (#3196)
- Add
CatBoostPruningCallback (#2734, thanks @tohmae!)
- Create common API for all NSGA-II crossover operations (#3221)
- Add a history of retried trial numbers in
Trial.system_attrs (#3223, thanks @belltailjp!)
- Convert all positional arguments to keyword-only (#3270, thanks @higucheese!)
- Stabilize
study.py (#3309)
- Add
targets and deprecate axis_order in optuna.visualization.matplotlib.plot_pareto_front (#3341, thanks @shu65!)
- Add
targets argument to plot_pareto_plont of plotly backend (#3495, thanks @TakuyaInoue-github!)
- Support
constraints_func in plot_pareto_front in matplotlib visualization (#3497, thanks @fukatani!)
- Calculate the feature importance with mean absolute SHAP values (#3507, thanks @liaison!)
- Make
GridSampler reproducible (#3527, thanks @gasin!)
- Replace
ValueError with warning in GridSearchSampler (#3545)
- Implement
callbacks argument of OptunaSearchCV (#3577)
- Add option to skip table creation to
RDBStorage (#3581)
- Add constraints option to
TPESampler (#3506)
- Add
skip_if_exists argument to enqueue_trial (#3629)
- Remove experimental from
plot_pareto_front (#3643)
- Add
popsize argument to CmaEsSampler (#3649)
- Add
seed argument for BoTorchSampler (#3756)
- Add
seed argument for SkoptSampler (#3791)
- Revert AllenNLP integration back to experimental (#3822)
- Remove abstractmethod decorator from
get_trial_id_from_study_id_trial_number (#3909)
Enhancements
- Add single distribution support to
BoTorchSampler (#2928)
- Speed up
import optuna (#3000)
- Fix
_contains of IntLogUniformDistribution (#3005)
- Render importance scores next to bars in
matplotlib.plot_param_importances (#3012, thanks @xadrianzetx!)
- Make default value of
verbose_eval NoneN forLightGBMTuner/LightGBMTunerCV` to avoid conflict (#3014, thanks @chezou!)
- Unify colormap of
plot_contour (#3017)
- Relax
FixedTrial and FrozenTrial allowing not-contained parameters during suggest_* (#3018)
- Raise errors if
optuna ask CLI receives --sampler-kwargs without --sampler (#3029)
- Remove
_get_removed_version_from_deprecated_version function (#3065, thanks @nuka137!)
- Reformat labels for small importance scores in
plotly.plot_param_importances (#3073, thanks @xadrianzetx!)
- Speed up Matplotlib backend
plot_contour using SciPy's spsolve (#3092)
- Remove updates in cached storage (#3120, thanks @shu65!)
- Reduce number of queries to fetch
directions, user_attrs and system_attrs of study summaries (#3108)
- Support
FloatDistribution across codebase (#3111, thanks @xadrianzetx!)
- Use
json.loads to decode pruner configuration loaded from environment variables (#3114)
- Show progress bar based on
timeout (#3115, thanks @xadrianzetx!)
- Support
IntDistribution across codebase (#3126, thanks @nyanhi!)
- Make progress bar available with n_jobs!=1 (#3138, thanks @masap!)
- Wrap
RedisStorage in CachedStorage (#3204, thanks @masap!)
- Use
functools.wraps in track_in_mlflow decorator (#3216)
- Make
RedisStorage fast when running multiple trials (#3262, thanks @masap!)
- Reduce database query result for
Study.ask() (#3274, thanks @masap!)
- Enable cache for
study.tell() (#3265, thanks @masap!)
- Warn if heartbeat is used with ask-and-tell (#3273)
- Make
optuna.study.get_all_study_summaries() of RedisStorage fast (#3278, thanks @masap!)
- Improve Ctrl-C interruption handling (#3374, thanks @CorentinNeovision!)
- Use same colormap among
plotly visualization methods (#3376)
- Make EDF plots handle trials with nonfinite values (#3435)
- Make logger message optional in
filter_nonfinite (#3438)
- Set
precision of sqlalchemy.Float in RDBStorage table definition (#3327)
- Accept
nan in trial.report (#3348, thanks @belldandyxtq!)
- Lazy import of alembic, sqlalchemy, and scipy (#3381)
- Unify pareto front (#3389, thanks @semiexp!)
- Make
set_trial_param() of RedisStorage faster (#3391, thanks @masap!)
- Make
_set_best_trial() of RedisStorage faster (#3392, thanks @masap!)
- Make
set_study_directions() of RedisStorage faster (#3393, thanks @masap!)
- Make optuna compatible with wandb sweep panels (#3403, thanks @captain-pool!)
- Change "#Trials" to "Trial" in
plot_slice, plot_pareto_front, and plot_optimization_history (#3449, thanks @dubey-anshuman!)
- Make contour plots handle trials with nonfinite values (#3451)
- Query studies for trials only once in EDF plots (#3460)
- Make Parallel-Coordinate plots handle trials with nonfinite values (#3471, thanks @divyanshugit!)
- Separate heartbeat functionality from
BaseStorage (#3475)
- Remove
torch.distributed calls from TorchDistributedTrial properties (#3490, thanks @nlgranger!)
- Remove the internal logic that calculates the interaction of two or more variables in fANOVA (#3543)
- Handle inf/-inf for
trial_values table in RDB (#3559)
- Add
intermediate_value_type column to represent inf/-inf on RDBStorage (#3564)
- Move
is_heartbeat_enabled from storage to heartbeat (#3596)
- Refactor
ImportanceEvaluators (#3597)
- Avoid maximum limit when MLflow saves information (#3651)
- Control metric decimal digits precision in
bayesmark benchmark report (#3693)
- Support
inf values for crowding distance (#3743)
- Normalize importance values (#3828)
Bug Fixes
- Add tests of
sample_relative and fix type of return values of SkoptSampler and PyCmaSampler (#2897)
- Fix
GridSampler with RetryFailedTrialCallback or enqueue_trial (#2946)
- Fix the type of
trial.values in MLflow integration (#2991)
- Fix to raise
ValueError for invalid q in DiscreteUniformDistribution (#3001)
- Do not call
trial.report during sanity check (#3002)
- Fix
matplotlib.plot_contour bug (#3046, thanks @IEP!)
- Handle
single distributions in fANOVA evaluator (#3085, thanks @xadrianzetx!)
- Fix bug of nondeterministic behavior of
TPESampler when group=True (#3187, thanks @xuzijian629!)
- Handle non-numerical params in
matplotlib.contour_plot (#3213, thanks @xadrianzetx!)
- Fix log scale axes padding in
matplotlib.contour_plot (#3218, thanks @xadrianzetx!)
- Handle
-inf and inf values in RDBStorage (#3238, thanks @xadrianzetx!)
- Skip limiting the value if it is
nan (#3286)
- Make TPE work with a categorical variable with different choice types (#3190, thanks @keisukefukuda!)
- Fix axis range issue in
matplotlib contour plot (#3249, thanks @harupy!)
- Allow
fail_state_trials show warning when heartbeat is enabled (#3301)
- Clip untransformed values sampled from int uniform distributions (#3319)
- Fix missing
user_attrs and system_attrs in study summaries (#3352)
- Fix objective scale in parallel coordinate of Matplotlib (#3369)
- Fix
matplotlib.plot_parallel_coordinate with log distributions (#3371)
- Fix parallel coordinate with missing value (#3373)
- Add utility to filter trials with
inf values from visualizations (#3395)
- Return the best trial number, not worst trial number by
best_index_ (#3410)
- Avoid using
px.colors.sequential.Blues that introduces pandas dependency (#3422)
- Fix
_is_reverse_scale (#3424)
- Import
COLOR_SCALE inside import util context (#3492)
- Remove
-v option of optuna study set-user-attr command (#3499, thanks @nyanhi!)
- Filter trials with nonfinite value in
optuna.visualization.plot_param_importances and optuna.visualization.matplotlib.plot_param_importance (#3500, thanks @takoika!)
- Fix
--verbose and --quiet options in CLI (#3532, thanks @nyanhi!)
- Replace
ValueError with RuntimeError in get_best_trial (#3541)
- Take the same search space as in
CategoricalDistribution by GridSampler (#3544)
- Fix
CategoricalDistribution with NaN (#3567)
- Fix NaN comparison in grid sampler (#3592)
- Fix bug in
IntersectionSearchSpace (#3666)
- Remove
trial_values records whose values are None (#3668)
- Fix PostgreSQL primary key unsorted problem (#3702, thanks @wattlebirdaz!)
- Raise error on NaN in
_constrained_dominates (#3738)
- Fix
inf-related issue on implementation of _calculate_nondomination_rank (#3739)
- Raise errors for NaN in constraint values (#3740)
- Fix
_calculate_weights such that it throws ValueError on invalid weights (#3742)
- Change warning for
axis_order of plot_pareto_front (#3802)
- Fix check for number of objective values (#3808)
- Raise
ValueError when waiting trial is told (#3814)
- Fix
Study.tell with invalid values (#3819)
- Fix infeasible case in NSGAII test (#3839)
Installation
- Support scikit-learn v1.0.0 (#3003)
- Pin
tensorflow and tensorflow-estimator versions to <2.7.0 (#3059)
- Add upper version constraint of PyTorchLightning (#3077)
- Pin
keras version to <2.7.0 (#3078)
- Remove version constraints of
tensorflow (#3084)
- Bump to
torch related packages (#3156)
- Use
pytorch-lightning>=1.5.0 (#3157)
- Remove testoutput from doctest of
mlflow integration (#3170)
- Restrict
nltk version (#3201)
- Add version constraints of
setuptools (#3207)
- Remove version constraint of
setuptools (#3231)
- Remove Sphinx version constraint (#3237)
- Drop TensorFlow support for Python 3.6 (#3296)
- Pin AllenNLP version (#3367)
- Skip run
fastai job on Python 3.6 (#3412)
- Avoid latest
click==8.1.0 that removed a deprecated feature (#3413)
- Avoid latest PyTorch lightning until integration is updated (#3417)
- Revert "Avoid latest
click==8.1.0 that removed a deprecated feature" (#3430)
- Partially support Python 3.10 (#3353)
- Clean up
setup.py (#3517)
- Remove duplicate requirements from
document section (#3613)
- Add a version constraint of cached-path (#3665)
- Relax version constraint of
fakeredis (#3905)
- Add version constraint for
typing_extensions to use ParamSpec (#3926)
Documentation
- Add note of the behavior when calling multiple
trial.report (#2980)
- Add note for DDP training of
pytorch-lightning (#2984)
- Add note to
OptunaSearchCV about direction (#3007)
- Clarify
n_trials in the docs (#3016, thanks @Rohan138!)
- Add a note to use pickle with different optuna versions (#3034)
- Unify the visualization docs (#3041, thanks @sidshrivastav!)
- Fix a grammatical error in FAQ doc (#3051, thanks @belldandyxtq!)
- Less ambiguous documentation for
optuna tell (#3052)
- Add example for
logging.set_verbosity (#3061, thanks @drumehiron!)
- Mention the tutorial of
002_configurations.py in the Trial API page (#3067, thanks @makkimaki!)
- Mention the tutorial of
003_efficient_optimization_algorithms.py in the Trial API page (#3068, thanks @makkimaki!)
- Add link from
set_user_attrs in Study to the user_attrs entry in Tutorial (#3069, thanks @MasahitoKumada!)
- Update description for missing samplers and pruners (#3087, thanks @masaaldosey!)
- Simplify the unit testing explanation (#3089)
- Fix range description in
suggest_float docstring (#3091, thanks @xadrianzetx!)
- Fix documentation for the package installation procedure on different OS (#3118, thanks @masap!)
- Add description of
ValueError and TypeErorr to Raises section of Trial.report (#3124, thanks @MasahitoKumada!)
- Add a note
logging_callback only works in single process situation (#3143)
- Correct
FrozenTrial's docstring (#3161)
- Promote to use of v3.0.0a0 in
README.md (#3167)
- Mention tutorial of callback for
Study.optimize from API page (#3171, thanks @xuzijian629!)
- Add reference to tutorial page in
study.enqueue_trial (#3172, thanks @knshnb!)
- Fix typo in specify_params (#3174, thanks @knshnb!)
- Guide to tutorial of Multi-objective Optimization in visualization tutorial (#3182, thanks @xuzijian629!)
- Add explanation about Parallelize Optimization at FAQ (#3186, thanks @MasahitoKumada!)
- Add order in tutorial (#3193, thanks @makinzm!)
- Fix inconsistency in
distributions documentation (#3222, thanks @xadrianzetx!)
- Add FAQ entry for heartbeat (#3229)
- Replace AUC with accuracy in docs (#3242)
- Fix
Raises section of FloatDistribution docstring (#3248, thanks @xadrianzetx!)
- Add
{Float,Int}Distribution to docs (#3252)
- Update explanation for metrics of
AllenNLPExecutor (#3253)
- Add missing cli methods to the list (#3268)
- Add docstring for property
DiscreteUniformDistribution.q (#3279)
- Add reference to tutorial page in CLI (#3267, thanks @tsukudamayo!)
- Carry over notes on
step behavior to new distributions (#3276)
- Correct the disable condition of
show_progress_bar (#3287)
- Add a document to lead FAQ and example of heartbeat (#3294)
- Add a note for
copy_study: it creates a copy regardless of its state (#3295)
- Add note to recommend Python 3.8 or later in documentation build with artifacts (#3312)
- Fix crossover references in
Raises doc section (#3315)
- Add reference to
QMCSampler in tutorial (#3320)
- Fix layout in tutorial (with workaround) (#3322)
- Scikit-learn required for
plot_param_importances (#3332, thanks @ll7!)
- Add a link to multi-objective tutorial from a pareto front page (#3339, thanks @kei-mo!)
- Add reference to tutorial page in visualization (#3340, thanks @Hiroyuki-01!)
- Mention tutorials of User-Defined Sampler/Pruner from the API reference pages (#3342, thanks @hppRC!)
- Add reference to saving/resuming study with RDB backend (#3345, thanks @Hiroyuki-01!)
- Fix a typo (#3360)
- Remove deprecated command
optuna study optimize in FAQ (#3364)
- Fix nit typo (#3380)
- Add see also section for
best_trial (#3396, thanks @divyanshugit!)
- Updates the tutorial page for re-use the best trial (#3398, thanks @divyanshugit!)
- Add explanation about
Study.best_trials in multi-objective optimization tutorial (#3443)
- Clean up exception docstrings (#3429)
- Revise docstring in MLFlow and WandB callbacks (#3477)
- Change the parameter name from
classifier to regressor in the code snippet of README.md (#3481)
- Add link to Minituna in
CONTRIBUTING.md (#3482)
- Fix
benchmarks/README.md for the bayesmark section (#3496)
- Mention
Study.stop as a criteria to stop creating trials in document (#3498, thanks @takoika!)
- Fix minor English errors in the docstring of
study.optimize (#3505)
- Add Python 3.10 in supported version in
README.md (#3508)
- Remove articles at the beginning of sentences in crossovers (#3509)
- Correct
FronzenTrial's docstring (#3514)
- Mention specify hyperparameter tutorial (#3515)
- Fix typo in MLFlow callback (#3533)
- Improve docstring of
GridSampler's seed option (#3568)
- Add the samplers comparison table (#3571)
- Replace
youtube.com with youtube-nocookie.com (#3590)
- Fix time complexity of the samplers comparison table (#3593)
- Remove
language from docs configuration (#3594)
- Add documentation of SHAP integration (#3623)
- Remove news entry on Optuna user survey (#3645)
- Introduce
optuna-fast-fanova (#3647)
- Add github discussions link (#3660)
- Fix a variable name of ask-and-tell tutorial (#3663)
- Clarify which trials are used for importance evaluators (#3707)
- Fix typo in
Study.optimize (#3720, thanks @29Takuya!)
- Update link to plotly's jupyterlab-support page (#3722, thanks @29Takuya!)
- Update
CONTRIBUTING.md (#3726)
- Remove "Edit on Github" button (#3777, thanks @cfkazu!)
- Remove duplicated period at the end of copyright (#3778)
- Add note for deprecation of
plot_pareto_front's axis_order (#3803)
- Describe the purpose of
prepare_study_with_trials (#3809)
- Fix a typo in docstring of
ShapleyImportanceEvaluator (#3810)
- Add a reference for MOTPE (#3838, thanks @y0z!)
- Minor fixes of sampler comparison table (#3850)
- Fix typo: Replace
trail with trial (#3861)
- Add
.. seealso:: in Study.get_trials and Study.trials (#3862, thanks @jmsykes83!)
- Add docstring of
TrialState.is_finished (#3869)
- Fix docstring in
FrozenTrial (#3872, thanks @wattlebirdaz!)
- Add note to explain when colormap reverses (#3873)
- Make
NSGAIISampler docs informative (#3880)
- Add note for
constant_liar with multi-objective function (#3881)
- Use
copybutton_prompt_text not to copy the bash prompt (#3882)
- Fix typo in
HyperbandPruner (#3894)
- Improve HyperBand docs (#3900)
- Mention reproducibility of
HyperBandPruner (#3901)
- Add a new note to mention unsupported GPU case for
CatBoostPruningCallback (#3903)
Examples
- Use
RetryFailedTrialCallback in pytorch_checkpoint example (https://github.com/optuna/optuna-examples/pull/59, thanks @xadrianzetx!)
- Add Python 3.9 to CI yaml files (https://github.com/optuna/optuna-examples/pull/61)
- Replace
suggest_uniform with suggest_float (https://github.com/optuna/optuna-examples/pull/63)
- Remove deprecated warning message in
lightgbm (https://github.com/optuna/optuna-examples/pull/64)
- Pin
tensorflow and tensorflow-estimator versions to <2.7.0 (https://github.com/optuna/optuna-examples/pull/66)
- Restrict upper version of
pytorch-lightning (https://github.com/optuna/optuna-examples/pull/67)
- Add an external resource to
README.md (https://github.com/optuna/optuna-examples/pull/68, thanks @solegalli!)
- Add pytorch-lightning DDP example (https://github.com/optuna/optuna-examples/pull/43, thanks @tohmae!)
- Install latest AllenNLP (https://github.com/optuna/optuna-examples/pull/73)
- Restrict
nltk version (https://github.com/optuna/optuna-examples/pull/75)
- Add version constraints of
setuptools (https://github.com/optuna/optuna-examples/pull/76)
- Remove constraint of
setuptools (https://github.com/optuna/optuna-examples/pull/79)
- Remove Python 3.6 from
haiku's CI (https://github.com/optuna/optuna-examples/pull/83)
- Apply
black 22.1.0 & run checks daily (https://github.com/optuna/optuna-examples/pull/84)
- Add
hiplot example (https://github.com/optuna/optuna-examples/pull/86)
- Stop running jobs using TF with Python3.6 (https://github.com/optuna/optuna-examples/pull/87)
- Pin AllenNLP version (https://github.com/optuna/optuna-examples/pull/89)
- Add Medium link (https://github.com/optuna/optuna-examples/pull/91)
- Use official
CatBoostPruningCallback (https://github.com/optuna/optuna-examples/pull/92)
- Stop running
fastai job on Python 3.6 (https://github.com/optuna/optuna-examples/pull/93)
- Specify Python version using
str in workflow files (https://github.com/optuna/optuna-examples/pull/95)
- Introduce upper version constraint of PyTorchLightning (https://github.com/optuna/optuna-examples/pull/96)
- Update
SimulatedAnnealingSampler to support FloatDistribution (https://github.com/optuna/optuna-examples/pull/97)
- Fix version of JAX (https://github.com/optuna/optuna-examples/pull/99)
- Remove constraints by #99 (https://github.com/optuna/optuna-examples/pull/100)
- Replace some methods in the
sklearn example (https://github.com/optuna/optuna-examples/pull/102, thanks @MasahitoKumada!)
- Add Python3.10 in
allennlp.yml (https://github.com/optuna/optuna-examples/pull/104)
- Remove numpy (https://github.com/optuna/optuna-examples/pull/105)
- Add python 3.10 to fastai CI (https://github.com/optuna/optuna-examples/pull/106)
- Add python 3.10 to non-integration examples CIs (https://github.com/optuna/optuna-examples/pull/107)
- Add python 3.10 to Hiplot CI (https://github.com/optuna/optuna-examples/pull/108)
- Add a comma to
visualization.yml (https://github.com/optuna/optuna-examples/pull/109)
- Rename WandB example to follow naming rules (https://github.com/optuna/optuna-examples/pull/110)
- Add scikit-learn version constraint for Dask-ML (https://github.com/optuna/optuna-examples/pull/112)
- Add python 3.10 to sklearn CI (https://github.com/optuna/optuna-examples/pull/113)
- Set version constraint of
protobuf in PyTorch Lightning example (https://github.com/optuna/optuna-examples/pull/116)
- Introduce stale bot (https://github.com/optuna/optuna-examples/pull/119)
- Use Hydra 1.2 syntax (https://github.com/optuna/optuna-examples/pull/122)
- Fix CI due to
thop (https://github.com/optuna/optuna-examples/pull/123)
- Hotfix
allennlp dependency (https://github.com/optuna/optuna-examples/pull/124)
- Remove unreferenced variable in
pytorch_simple.py (https://github.com/optuna/optuna-examples/pull/125)
- set
OMPI_MCA_rmaps_base_oversubscribe=yes before mpirun (https://github.com/optuna/optuna-examples/pull/126)
- Add python 3.10 to
python-version (https://github.com/optuna/optuna-examples/pull/127)
- Remove upper version constraint of
sklearn (https://github.com/optuna/optuna-examples/pull/128)
- Move
catboost integration line to integration section from pruning section (https://github.com/optuna/optuna-examples/pull/129)
- Simplify
skimage example (https://github.com/optuna/optuna-examples/pull/130)
- Remove deprecated warning in PyTorch Lightning example (https://github.com/optuna/optuna-examples/pull/131)
- Resolve TODO task in ray example (https://github.com/optuna/optuna-examples/pull/132)
- Remove version constraint of
cached-path (https://github.com/optuna/optuna-examples/pull/133)
Tests
- Add test case of samplers for conditional objective function (#2904)
- Test int distributions with default step (#2924)
- Be aware of trial preparation when checking heartbeat interval (#2982)
- Simplify the DDP model definition in the test of
pytorch-lightning (#2983)
- Wrap data with
np.asarray in lightgbm test (#2997)
- Patch calls to deprecated
suggest APIs across codebase (#3027, thanks @xadrianzetx!)
- Make
return_cvbooster of LightGBMTuner consistent to the original value (#3070, thanks @abatomunkuev!)
- Fix
parametrize_sampler (#3080)
- Fix verbosity for
tests/integration_tests/lightgbm_tuner_tests/test_optimize.py (#3086, thanks @nyanhi!)
- Generalize empty search space test case to all hyperparameter importance evaluators (#3096, thanks @xadrianzetx!)
- Check if texts in legend by order agnostic way (#3103)
- Add tests for axis scales to
matplotlib.plot_slice (#3121)
- Add tests for transformer with upper bound parameter (#3163)
- Add tests in
visualization_tests/matplotlib_tests/test_slice.py (#3175, thanks @keisukefukuda!)
- Add test case of the value in optimization history with matplotlib (#3176, thanks @TakuyaInoue-github!)
- Add tests for generated plots of
matplotlib.plot_edf (#3178, thanks @makinzm!)
- Improve pareto front figure tests for matplotlib (#3183, thanks @akawashiro!)
- Add tests for generated plots of
plot_edf (#3188, thanks @makinzm!)
- Match contour tests between Plotly and Matplotlib (#3192, thanks @belldandyxtq!)
- Implement missing
matplotlib.contour_plot test (#3232, thanks @xadrianzetx!)
- Unify the validation function of edf value between visualization backends (#3233)
- Add test for default grace period (#3263, thanks @masap!)
- Add the missing tests of Plotly's
plot_parallel_coordinate (#3266, thanks @MasahitoKumada!)
- Switch function order progbar tests (#3280, thanks @BasLaa!)
- Add plot value tests to
matplotlib_tests/test_param_importances (#3180, thanks @belldandyxtq!)
- Make tests of
plot_optimization_history methods consistent (#3234)
- Add integration test for
RedisStorage (#3258, thanks @masap!)
- Change the order of arguments in the
catalyst integration test (#3308)
- Cleanup
MLflowCallback tests (#3378)
- Test serialize/deserialize storage on parametrized conditions (#3407)
- Add tests for parameter of 'None' for TPE (#3447)
- Improve
matplotlib parallel coordinate test (#3368)
- Save figures for all
matplotlib tests (#3414, thanks @divyanshugit!)
- Add
inf test to intermediate values test (#3466)
- Add test cases for
test_storages.py (#3480)
- Improve the tests of
optuna.visualization.plot_pareto_front (#3546)
- Move heartbeat-related tests in
test_storages.py to another file (#3553)
- Use
seed method of np.random.RandomState for reseeding and fix test_reseed_rng (#3569)
- Refactor
test_get_observation_pairs (#3574)
- Add tests for
inf/nan objectives for ShapleyImportanceEvaluator (#3576)
- Add deprecated warning test to the multi-objective sampler test file (#3601)
- Simplify multi-objective TPE tests (#3653)
- Add edge cases to multi-objective TPE tests (#3662)
- Remove tests on
TypeError (#3667)
- Add edge cases to the tests of the parzen estimator (#3673)
- Add tests for
_constrained_dominates (#3683)
- Refactor tests of constrained TPE (#3689)
- Add
inf and NaN tests for test_constraints_func (#3690)
- Fix calling storage API in study tests (#3695, thanks @wattlebirdaz!)
- DRY
test_frozen.py (#3696)
- Unify the tests of
plot_contours (#3701)
- Add test cases for crossovers of NSGAII (#3705)
- Enhance the tests of
NSGAIISampler._crowding_distance_sort (#3706)
- Unify edf test files (#3730)
- Fix
test_calculate_weights_below (#3741)
- Refactor
test_intermediate_plot.py (#3745)
- Test samplers are reproducible (#3757)
- Add tests for
_dominates function (#3764)
- DRY importance tests (#3785)
- Move tests for
create_trial (#3794)
- Remove
with_c_d option from prepare_study_with_trials (#3799)
- Use
DeterministicRelativeSampler in test_trial.py (#3807)
- Add tests for
_fast_non_dominated_sort (#3686)
- Unify slice plot tests (#3784)
- Unify the tests of
plot_parallel_coordinates (#3800)
- Unify optimization history tests (#3806)
- Suppress warnings in tests for
multi_objective module (#3911)
- Remove
warnings: UserWarning from tests/visualization_tests/test_utils.py (#3919, thanks @jmsykes83!)
Code Fixes
- Add test case of samplers for conditional objective function (#2904)
- Fix #2949, remove
BaseStudy (#2986, thanks @twsl!)
- Use
optuna.load_study in optuna ask CLI to omit direction/directions option (#2989)
- Fix typo in
Trial warning message (#3008, thanks @xadrianzetx!)
- Replaces boston dataset with california housing dataset (#3011, thanks @avats-dev!)
- Fix deprecation version of
suggest APIs (#3054, thanks @xadrianzetx!)
- Add
remove_version to the missing @deprecated argument (#3064, thanks @nuka137!)
- Add example of
optuna.logging.get_verbosity (#3066, thanks @MasahitoKumada!)
- Support
{Float|Int}Distribution in NSGA-II crossover operators (#3139, thanks @xadrianzetx!)
- Black fix (#3147)
- Switch to
FloatDistribution (#3166, thanks @xadrianzetx!)
- Remove
deprecated decorator of the feature of n_jobs (#3173, thanks @MasahitoKumada!)
- Fix black and blackdoc errors (#3260, thanks @masap!)
- Remove experimental label from
MaxTrialsCallback (#3261, thanks @knshnb!)
- Remove redundant
_check_trial_id (#3264, thanks @masap!)
- Make existing int/float distributions wrapper of
{Int,Float}Distribution (#3244)
- Switch to
IntDistribution (#3181, thanks @nyanhi!)
- Fix type hints for Python 3.8 (#3240)
- Remove
UniformDistribution, LogUniformDistribution and DiscreteUniformDistribution code paths (#3275)
- Merge
set_trial_state() and set_trial_values() into one function (#3323, thanks @masap!)
- Follow up for
{Float, Int}Distributions (#3337, thanks @nyanhi!)
- Move the
get_trial_xxx abstract functions to base (#3338, thanks @belldandyxtq!)
- Update type hints of
states (#3359, thanks @BasLaa!)
- Remove unused function from
RedisStorage (#3394, thanks @masap!)
- Remove unnecessary string concatenation (#3406)
- Follow coding style and fix typos in
tests/integration_tests (#3408)
- Fix log message formatting in
filter_nonfinite (#3436)
- Add
RetryFailedTrialCallback to optuna.storages.* (#3441)
- Unify
fail_stale_trials in each storage implementation (#3442, thanks @knshnb!)
- Ignore incomplete trials in
matplotlib.plot_parallel_coordinate (#3415)
- Update warning message and add a test when a trial fails with exception (#3454)
- Remove old distributions from NSGA-II sampler (#3459)
- Remove duplicated DB access in
_log_completed_trial (#3551)
- Reduce the number of
copy.deepcopy() calls in importance module (#3554)
- Remove duplicated
check_trial_is_updatable (#3557)
- Replace
optuna.testing.integration.create_running_trial with study.ask (#3562)
- Refactor
test_get_observation_pairs (#3574)
- Update label of feasible trials if
constraints_func is specified (#3587)
- Replace unused variable name with underscore (#3588)
- Enable
no-implicit-optional for mypy (#3599, thanks @harupy!)
- Enable
warn_redundant_casts for mypy (#3602, thanks @harupy!)
- Refactor the type of value of
TrialIntermediateValueModel (#3603)
- Fix broken
mypy checks of Alembic's get_current_head() method (#3608)
- Move heartbeat-related thread operation in
_optimize.py to _heartbeat.py (#3609)
- Sort dependencies by name (#3614)
- Add typehint for deprecated and experimental (#3575)
- Remove useless object inheritance (#3628, thanks @harupy!)
- Remove useless
except clauses (#3632, thanks @harupy!)
- Rename
optuna.testing.integration with optuna.testing.pruner (#3638)
- Cosmetic fix in Optuna CLI (#3641)
- Enable
strict_equality for mypy #3579 (#3648, thanks @wattlebirdaz!)
- Make file names in testing consistent with
optuna module (#3657)
- Remove the implementation of
read_trials_from_remote_storage in the all storages apart from CachedStorage (#3659)
- Remove unnecessary deep copy in Redis storage (#3672, thanks @wattlebirdaz!)
- Workaround
mypy bug (#3679)
- Unify
plot_contours (#3682)
- Remove
storage.get_all_study_summaries(include_best_trial: bool) (#3697, thanks @wattlebirdaz!)
- Unify the logic of edf functions (#3698)
- Unify the logic of
plot_param_importances functions (#3700)
- Enable
disallow_untyped_calls for mypy (#3704, thanks @29Takuya!)
- Use
get_trials with states argument to filter trials depending on trial state (#3708)
- Return Python's native float values (#3714)
- Simplify
bayesmark benchmark report rendering (#3725)
- Unify the logic of intermediate plot (#3731)
- Unify the logic of slice plot (#3732)
- Unify the logic of
plot_parallel_coordinates (#3734)
- Unify implementation of
plot_optimization_history between plotly and matplotlib (#3736)
- Extract
fail_objective and pruned_objective for tests (#3737)
- Remove deprecated storage functions (#3744, thanks @29Takuya!)
- Remove unnecessary optionals from
visualization/_pareto_front.py (#3752)
- Change types inside
_ParetoInfoType (#3753)
- Refactor pareto front (#3754)
- Use
_ContourInfo to plot in plot_contour (#3755)
- Follow up #3465 (#3763)
- Refactor importances plot (#3765)
- Remove
no_trials option of prepare_study_with_trials (#3766)
- Follow the coding style of comments in
plot_contour files (#3767)
- Raise
ValueError for invalid returned type of target in _filter_nonfinite (#3768)
- Fix value error condition in
plot_contour (#3769)
- DRY constraints in
Sampler.after_trial (#3775)
- DRY
stop_objective (#3786)
- Refactor non-exist param test in
plot_contour test (#3787)
- Remove
less_than_two and more_than_three options from prepare_study_with_trials (#3789)
- Fix return value's type of
_get_node_value (#3818)
- Remove unused
type: ignore (#3832)
- Fix typos and remove unused argument in
QMCSampler (#3837)
- Unify tests for
plot_param_importances (#3760)
- Refactor
test_pareto_front (#3798)
- Remove duplicated definition of
CategoricalChoiceType from optuna.distributions (#3846)
- Revert most of changes by 3651 (#3848)
- Attach abstractmethod decorator to
BaseStorage.get_trial_id_from_study_id_trial_number (#3870, thanks @wattlebirdaz!)
- Refactor
BaseStorage.get_best_trial (#3871, thanks @wattlebirdaz!)
- Simplify
IntersectionSearchSpace.calculate (#3887)
- Replace
q with step in private function and warning message (#3913)
- Reduce warnings in storage tests (#3917)
- Reduce trivial warning messages from
tests/sampler_tests (#3921)
Continuous Integration
- Install
botorch to CI jobs on mac (#2988)
- Use libomp 11.1.0 for Mac (#3024)
- Run
mac-tests CI at a scheduled time (#3028)
- Set concurrency to github workflows (#3095)
- Skip CLI tests when calculating the coverage (#3097)
- Migrate
mypy version to 0.910 (#3123)
- Avoid installing the latest MLfow to prevent doctests from failing (#3135)
- Use python 3.8 for CI and docker (#3026)
- Add performance benchmarks using
kurobako (#3155)
- Use Python 3.7 in checks CI job (#3239)
- Add performance benchmarks using
bayesmark (#3354)
- Fix speed benchmarks (#3362)
- Pin
setuptools (#3427)
- Introduce the benchmark for multi-objectives samplers (#3271, thanks @drumehiron!)
- Use
coverage directly (#3347, thanks @higucheese!)
- Add WFG benchmark test (#3349, thanks @kei-mo!)
- Add workflow to use
reviewdog (#3357)
- Add NASBench201 from NASLib (#3465)
- Fix speed benchmarks CI (#3470)
- Support PyTorch 1.11.0 (#3510)
- Install 3rd party libraries in CI for lint (#3580)
- Make
bayesmark benchmark results comparable to kurobako (#3584)
- Restore
virtualenv for benchmark extras (#3585)
- Use
protobuf<4.0.0 to resolve Sphinx CI error (#3591)
- Unpin
protobuf (#3598, thanks @harupy!)
- Extract MPI tests from integration CI as independent CI (#3606)
- Enable
warn_unused_ignores for mypy (#3627, thanks @harupy!)
- Add
onnx and version constrained protobuf to document dependencies (#3658)
- Add
mo-kurobako benchmark to CI (#3691)
- Enable mypy's strict configs (#3710)
- Run visual regression tests to find regression bugs of visualization module (#3721)
- Remove downloading old
libomp for mac tests (#3728)
- Match Python versions between
bayesmark CI jobs (#3750)
- Set
OMPI_MCA_rmaps_base_oversubscribe=yes before mpirun (#3758)
- Add
budget option to benchmarks (#3774)
- Add
n_concurrency option to benchmarks (#3776)
- Use
n-runs instead of repeat to represent the number of studies in the bayesmark benchmark (#3780)
- Fix type hints for
mypy 0.971 (#3797)
- Pin scipy to avoid the CI failure (#3834)
- Extract float value from tensor for
trial.report in PyTorchLightningPruningCallback (#3842)
Other
- Bump up version to 2.11.0dev (#2976)
- Add roadmap news to
README.md (#2999)
- Bump up version number to 3.0.0a1.dev (#3006)
- Add Python 3.9 to
tox.ini (#3025)
- Fix version number to 3.0.0a0 (#3140)
- Bump up version to v3.0.0a1.dev (#3142)
- Introduce a form to make TODOs explicit when creating issues (#3169)
- Bump up version to
v3.0.0b0.dev (#3289)
- Add description field for
question-and-help-support (#3305)
- Update README to inform
v3.0.0a2 (#3314)
- Add Optuna-related URLs for PyPi (#3355, thanks @andriyor!)
- Bump Optuna to
v3.0.0-b0 (#3458)
- Bump up version to v3.0.0b1.dev (#3457)
- Fix
kurobako benchmark code to run it locally (#3468)
- Fix label of issue template (#3493)
- Improve issue templates (#3536)
- Hotfix for
fakeredis 1.7.4 release (#3549)
- Remove the version constraint of
fakeredis (#3561)
- Relax version constraint of
fakeredis (#3607)
- Shorten the durations of the stale bot for PRs (#3611)
- Clarify the criteria to assign reviewers in the PR template (#3619)
- Bump up version number to v3.0.0rc0.dev (#3621)
- Make
tox.ini consistent with checking (#3654)
- Avoid to stale description-checked issues (#3816)
- Bump up version to v3.0.0.dev (#3852)
- Bump up version to v3.0.0 (#3933)
Thanks to All the Contributors!
This release was made possible by the authors and the people who participated in the reviews and discussions.
@29Takuya, @BasLaa, @CorentinNeovision, @Crissman, @HideakiImamura, @Hiroyuki-01, @IEP, @MasahitoKumada, @Rohan138, @TakuyaInoue-github, @abatomunkuev, @akawashiro, @andriyor, @avats-dev, @belldandyxtq, @belltailjp, @c-bata, @captain-pool, @cfkazu, @chezou, @contramundum53, @divyanshugit, @drumehiron, @dubey-anshuman, @fukatani, @g-votte, @gasin, @harupy, @higucheese, @himkt, @hppRC, @hvy, @jmsykes83, @kasparthommen, @kei-mo, @keisuke-umezawa, @keisukefukuda, @knshnb, @kstoneriv3, @liaison, @ll7, @makinzm, @makkimaki, @masaaldosey, @masap, @nlgranger, @not522, @nuka137, @nyanhi, @nzw0301, @semiexp, @shu65, @sidshrivastav, @sile, @solegalli, @takoika, @tohmae, @toshihikoyanase, @tsukudamayo, @tupui, @twsl, @wattlebirdaz, @xadrianzetx, @xuzijian629, @y0z, @yoshinobc, @ytsmiling
Source code(tar.gz)
Source code(zip)
























 |
|  |
|

 |
|  |
|





 |
|  |
|


 |
|  |
|  |
|

 |
|