Large Steps in Inverse Rendering of Geometry
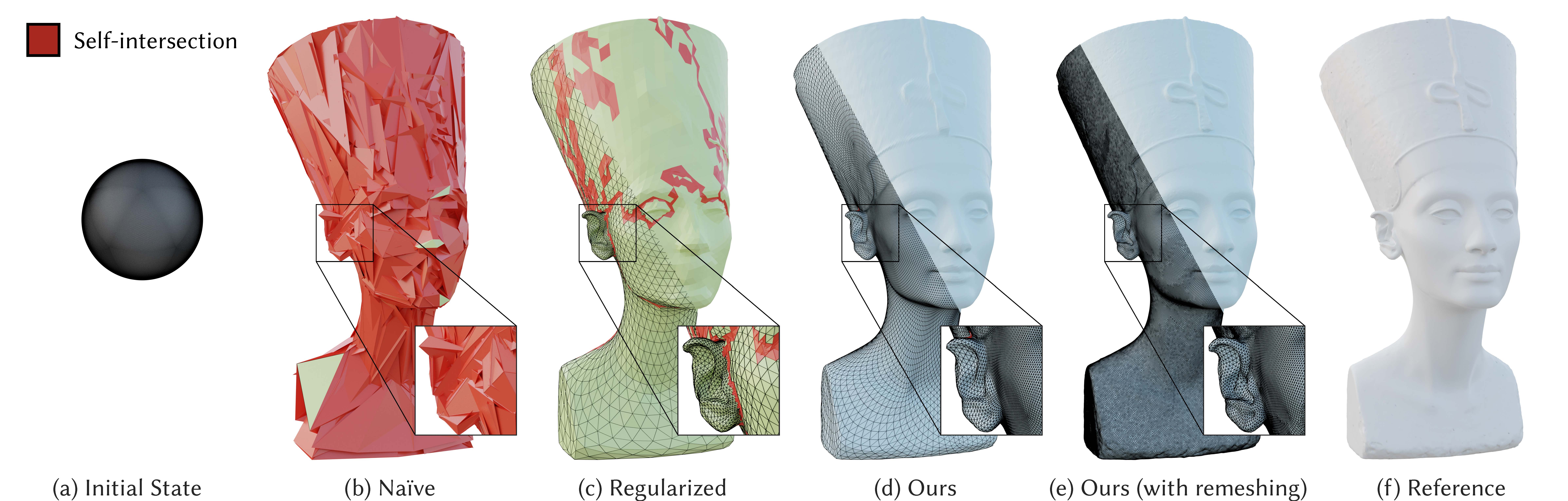
ACM Transactions on Graphics (Proceedings of SIGGRAPH Asia), December 2021.
Baptiste Nicolet · Alec Jacobson · Wenzel Jakob
Table of Contents
Installation
This repository contains both the operators needed to use our parameterization of vertex positions of meshes as well as the code for the experiments we show in the paper.
Parameterization package installation
If you are only interested in using our parameterization in an existing (PyTorch based) pipeline, we have made it available to install via pip. However, it depends on cupy and scikit-sparse, which need to be installed manually beforehand. We first need to install the suitesparse dependency.
# Ubuntu/Debian
apt install libsuitesparse-dev
# Fedora
yum install suitesparse-devel
# Arch linux
pacman -S suitesparse
# Mac OS X
brew install suite-sparse
Then install the python dependencies via pip:
pip install cupy-cudaXXX # Adjust this to your CUDA version, following https://docs.cupy.dev/en/stable/install.html#installing-cupy
pip install scikit-sparse
Then, install our package:
pip install largesteps
This will install the largesteps module. This only contains the parameterization logic implemented as a PyTorch custom operator. See the tutorial for an example use case.
Cloning the repository
Otherwise, if you want to reproduce the experiments from the paper, you can clone this repo and install the module locally. Make sure you have installed the cupy and scikit-sparse dependencies mentioned above before.
git clone --recursive [email protected]:rgl-epfl/large-steps-pytorch.git
cd large-steps-pytorch
pip install .
The experiments in this repository depend on PyTorch. Please follow instructions on the PyTorch website to install it.
To install nvdiffrast and the Botsch-Kobbelt remesher, which are provided as submodules, please run the setup_dependencies.sh script.
To install the other dependencies needed to run the experiments, also run:
pip install -r requirements.txt
nvdiffrast requires using g++ to compile some PyTorch extensions, make sure this is your default compiler:
export CC=gcc && CXX=g++
Rendering the figures will also require installing blender. You can specify the name of the blender executable you wish to use in scripts/constants.py
Downloading the scenes
The scenes for the experiments can be downloaded here. Please extract the archive at the toplevel of this repository.
Parameterization
In a nutshell, our parameterization can be obtained in just a few lines:
# Given tensors v and f containing vertex positions and faces
from largesteps.geometry import laplacian_uniform, compute_matrix
from largesteps.parameterize import to_differential, from_differential
L = laplacian_uniform(v, f)
M = compute_matrix(L, lambda_=10)
u = to_differential(v, M)
compute_matrix returns the parameterization matrix M = I + λL. This function takes another parameter, alpha, which leads to a slightly different, but equivalent, formula for the matrix: M = (1-α)I + αL, with α ∈ [0,1[. With this formula, the scale of the matrix M has the same order of magnitude regardless of α.
M = compute_matrix(L, alpha=0.9)
Then, vertex coordinates can be retrieved as:
v = from_differential(u, M, method='Cholesky')
This will in practice perform a cache lookup for a solver associated to the matrix M (and instantiate one if not found) and solve the linear system Mv = u. Further calls to from_differential with the same matrix will use the solver stored in the cache. Since this operation is implemented as a differentiable PyTorch operation, there is nothing more to be done to optimize this parameterization.
Running the experiments
You can then run the experiments in the figures folder, in which each subfolder corresponds to a figure in the paper, and contains two files:
generate_data.py: contains the script to run the experiment and write the output to the directory specified inscripts/constants.pyfigure.ipynb: contains the script generating the figure, assuminggenerate_data.pyhas been run before and the output written to the directory specified inscripts/constants.py
We provide the scripts for the following figures:
- Fig. 1 ->
teaser - Fig. 3 ->
multiscale - Fig. 5 ->
remeshing - Fig. 6 ->
reg_fail - Fig. 7 ->
comparison - Fig. 8 ->
viewpoints - Fig. 9 ->
influence
Repository structure
The largesteps folder contains the parameterization module made available via pip. It contains:
geometry.py: contains the laplacian matrix computation.optimize.py: contains theAdamUniformoptimizer implementationparameterize.py: contains the actual parameterization code, implemented as ato_differentialandfrom_differentialfunction.solvers.py: contains the Cholesky and conjugate gradients solvers used to convert parameterized coordinates back to vertex coordinates.
Other functions used for the experiments are included in the scripts folder:
blender_render.py: utility script to render meshes inside blenderconstants.py: contains paths to different useful folders (scenes, remesher, etc.)geometry.py: utility geometry functions (normals computation, edge length, etc.)io_ply.py: PLY mesh file loadingload_xml.py: XML scene file loadingmain.py: contains the main optimization functionpreamble.py: utility scipt to a import redundant modules for the figuresrender.py: contains the rendering logic, usingnvdiffrast
License
This code is provided under a 3-clause BSD license that can be found in the LICENSE file. By using, distributing, or contributing to this project, you agree to the terms and conditions of this license.
Citation
If you use this code for academic research, please cite our method using the following BibTeX entry:
@article{Nicolet2021Large,
author = "Nicolet, Baptiste and Jacobson, Alec and Jakob, Wenzel",
title = "Large Steps in Inverse Rendering of Geometry",
journal = "ACM Transactions on Graphics (Proceedings of SIGGRAPH Asia)",
volume = "40",
number = "6",
year = "2021",
month = dec,
doi = "10.1145/3478513.3480501",
url = "https://rgl.epfl.ch/publications/Nicolet2021Large"
}
Acknowledgments
The authors would like to thank Delio Vicini for early discussions about this project, Silvia Sellán for sharing her remeshing implementation and help for the figures, as well as Hsueh-Ti Derek Liu for his advice in making the figures. Also, thanks to Miguel Crespo for making this README template.


