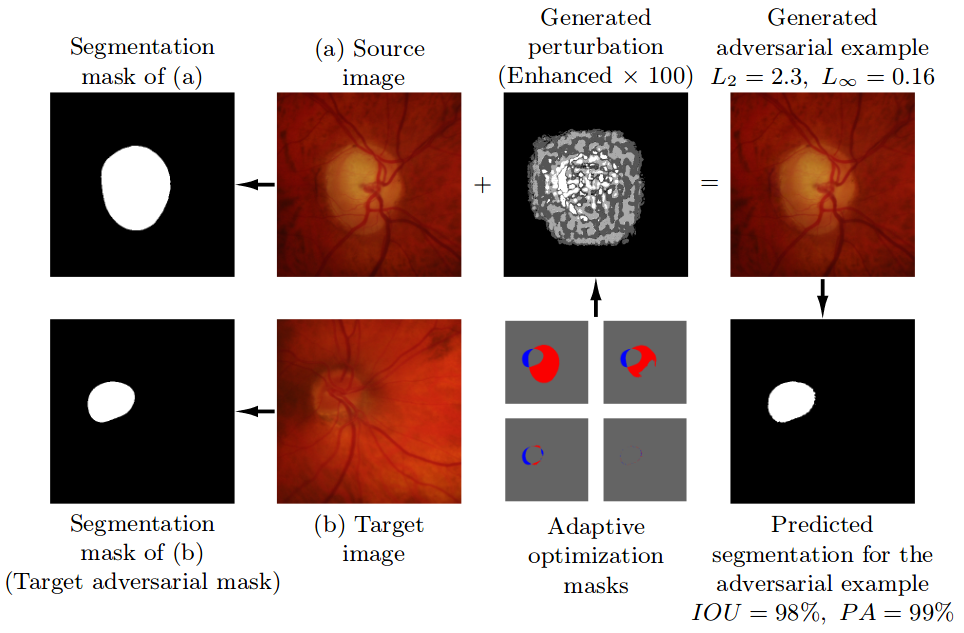
Adaptive Segmentation Mask Attack
This repository contains the implementation of the Adaptive Segmentation Mask Attack (ASMA), a targeted adversarial example generation method for deep learning segmentation models. This attack was proposed in the paper "Impact of Adversarial Examples on Deep Learning Models for Biomedical Image Segmentation." published in the 22nd International Conference on Medical Image Computing and Computer Assisted Intervention, MICCAI-2019. (Link to the paper)
General Information
This repository is organized as follows:
-
Code - src/ folder contains necessary python files to perform the attack and calculate various stats (i.e., correctness and modification)
-
Data - data/ folder contains a couple of examples for testing purposes. The data we used in this study can be taken from [1].
-
Model - Example model used in this repository can be downloaded from https://www.dropbox.com/s/6ziz7s070kkaexp/eye_pretrained_model.pt . helper_functions.py contains a function to load this file and main.py contains an exaple that uses this model.
Frequently Asked Questions (FAQ)
-
How can I run the demo?
1- Download the model from https://www.dropbox.com/s/6ziz7s070kkaexp/eye_pretrained_model.pt
2- Create a folder called model on the same level as data and src, put the model under this (model) folder.
3- Run main.py.
-
Would this attack work in multi-class segmentation models?
Yes, given that you provide a proper target mask, model etc.
-
Does the code require any modifications in order to make it work for multi-class segmentation models?
No (probably, depending on your model/input). At least the attack itself (adaptive_attack.py) should not require major modifications on its logic.
Citation
If you find the code in this repository useful for your research, consider citing our paper. Also, feel free to use any visuals available here.
@inproceedings{ozbulak2019impact,
title={Impact of Adversarial Examples on Deep Learning Models for Biomedical Image Segmentation},
author={Ozbulak, Utku and Van Messem, Arnout and De Neve, Wesley},
booktitle={International Conference on Medical Image Computing and Computer-Assisted Intervention},
pages={300--308},
year={2019},
organization={Springer}
}
Requirements
python > 3.5
torch >= 0.4.0
torchvision >= 0.1.9
numpy >= 1.13.0
PIL >= 1.1.7
References
[1] Pena-Betancor C., Gonzalez-Hernandez M., Fumero-Batista F., Sigut J., Medina-Mesa E., Alayon S., Gonzalez M. Estimation of the relative amount of hemoglobin in the cup and neuroretinal rim using stereoscopic color fundus images.
[2] Ronneberger, O., Fischer, P., Brox, T. U-Net: Convolutional networks for biomedical image segmentation.