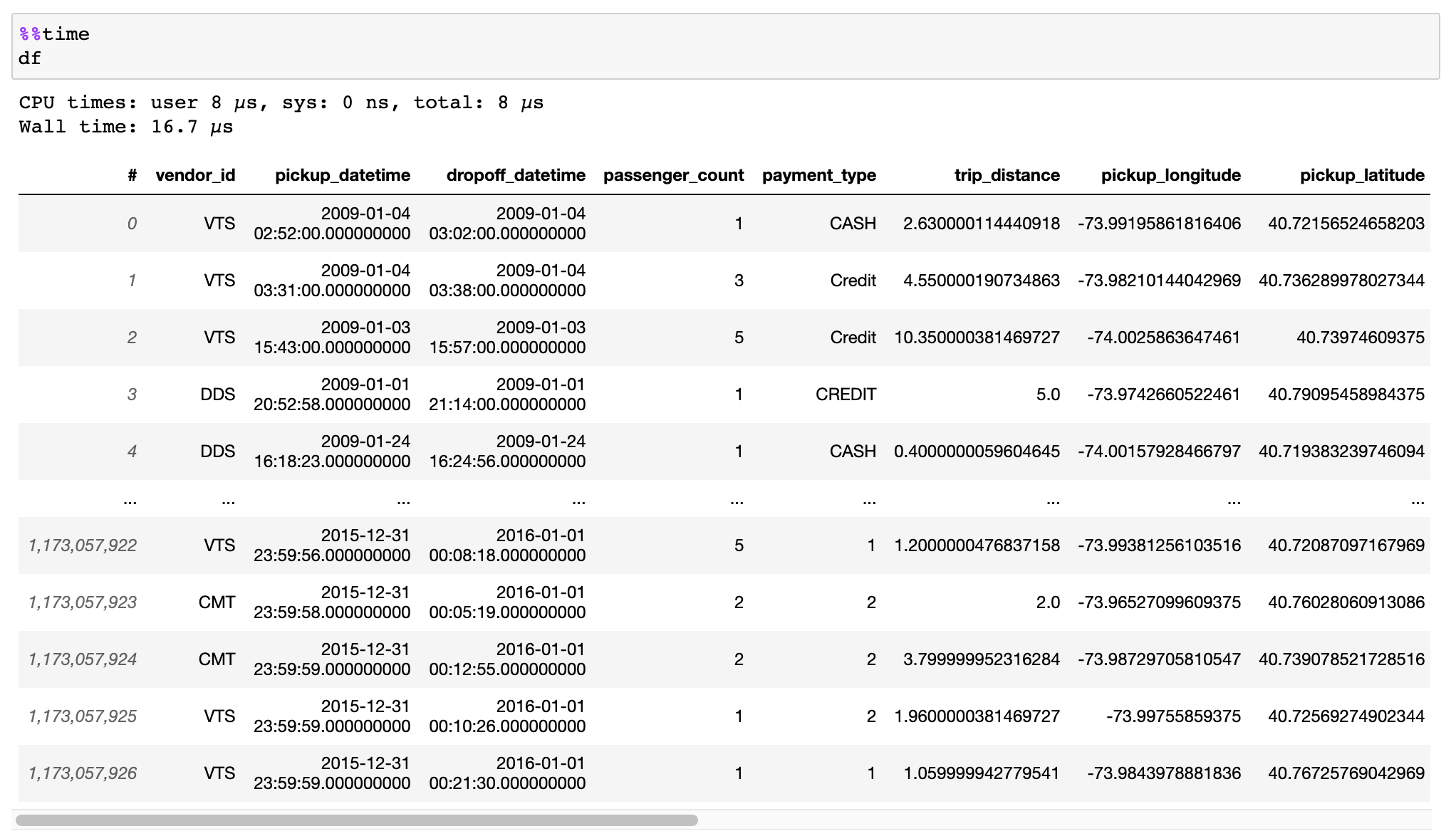
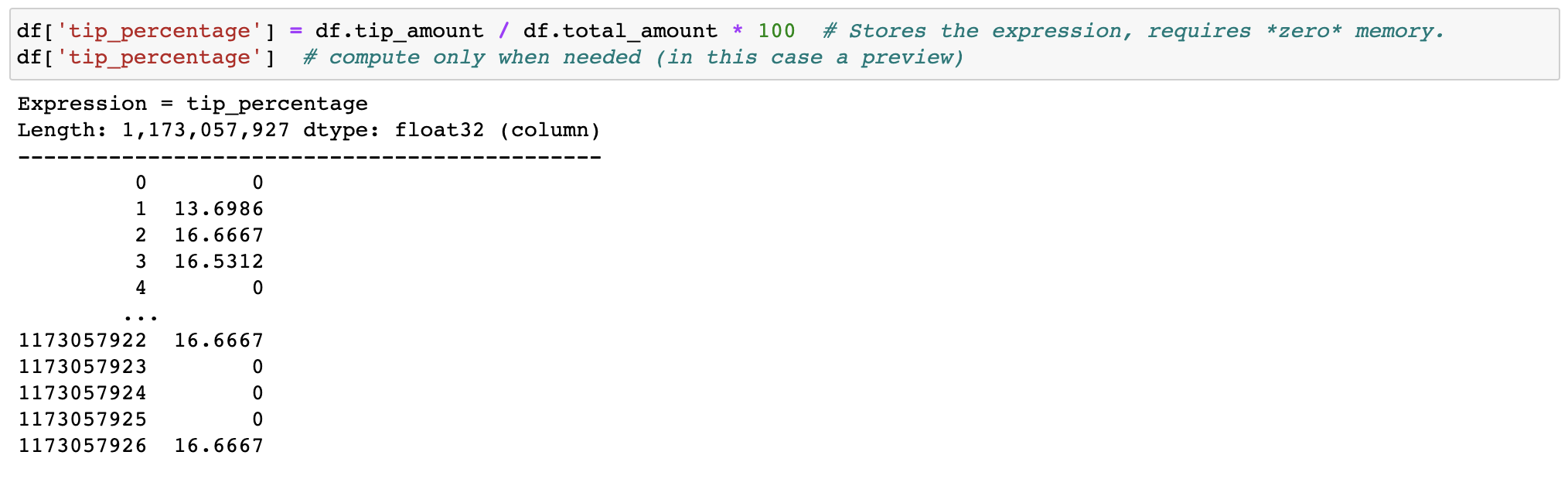
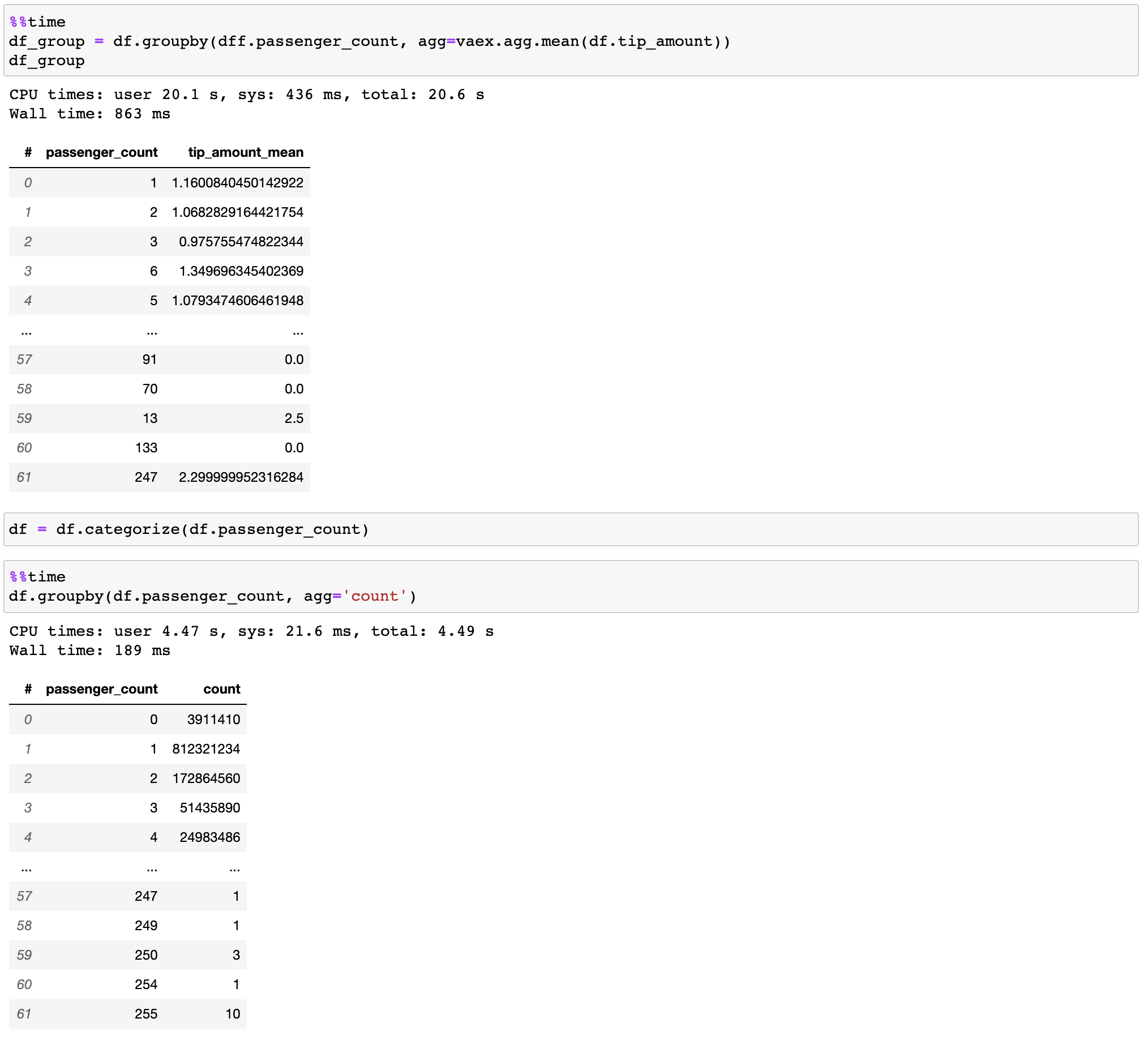
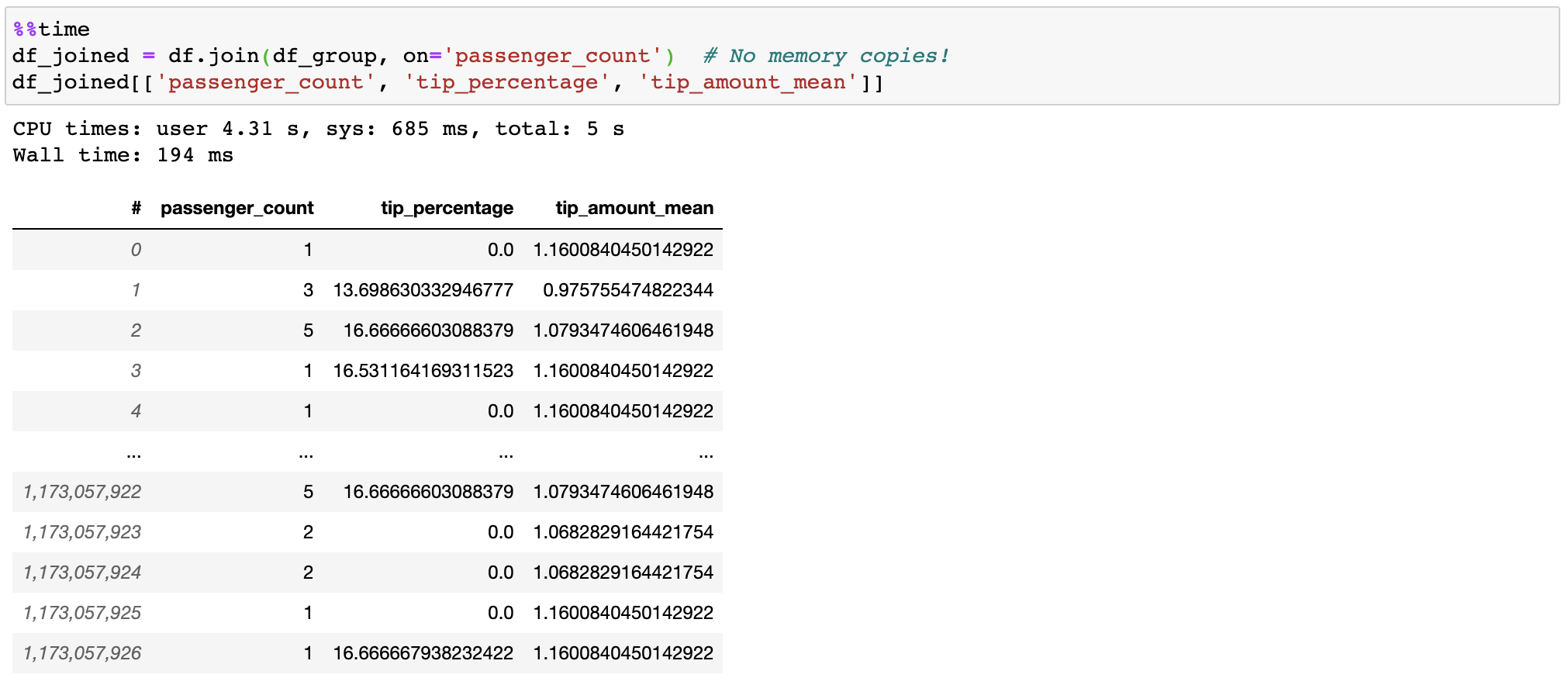
I have attempted to utilize vaex, but have been unsuccessful. Reason unknown.
I am using the most recent version of anaconda 3. I just uninstalled and reinstalled the most recent version as I was experiencing this same problem and figured a fresh install may help to clear the issue. The same error is appearing. I have copied and pasted the console here.
Python 3.8.5 (default, Sep 3 2020, 21:29:08) [MSC v.1916 64 bit (AMD64)]
Type "copyright", "credits" or "license" for more information.
IPython 7.19.0 -- An enhanced Interactive Python.
conda install -c conda-forge vaex
Collecting package metadata (current_repodata.json): ...working... done
Solving environment: ...working... failed with initial frozen solve. Retrying with flexible solve.
Solving environment: ...working... failed with repodata from current_repodata.json, will retry with next repodata source.
Collecting package metadata (repodata.json): ...working... done
Solving environment: ...working... done
Package Plan
environment location: C:\Users\sutei\anaconda3
added / updated specs:
- vaex
The following packages will be downloaded:
package | build
---------------------------|-----------------
abseil-cpp-20200225.2 | ha925a31_2 1.9 MB conda-forge
aplus-0.11.0 | py_1 6 KB conda-forge
arrow-cpp-2.0.0 |py38h647f3f1_6_cpu 13.4 MB conda-forge
aws-c-common-0.4.59 | h8ffe710_1 151 KB conda-forge
aws-c-event-stream-0.1.6 | hb4e73fc_6 26 KB conda-forge
aws-checksums-0.1.10 | h6f0a1a5_0 51 KB conda-forge
aws-sdk-cpp-1.8.70 | he2782d2_1 2.9 MB conda-forge
boto3-1.17.19 | pyhd8ed1ab_0 70 KB conda-forge
botocore-1.20.19 | pyhd8ed1ab_0 4.5 MB conda-forge
bqplot-0.12.20 | pyhd8ed1ab_0 1.0 MB conda-forge
branca-0.4.2 | pyhd8ed1ab_0 26 KB conda-forge
brotli-1.0.9 | h0e60522_4 882 KB conda-forge
c-ares-1.17.1 | h8ffe710_0 109 KB conda-forge
cachetools-4.2.1 | pyhd8ed1ab_0 13 KB conda-forge
conda-4.9.2 | py38haa244fe_0 3.1 MB conda-forge
geos-3.9.1 | h39d44d4_2 1.1 MB conda-forge
gflags-2.2.2 | ha925a31_1004 80 KB conda-forge
glog-0.4.0 | h0174b99_3 83 KB conda-forge
grpc-cpp-1.33.2 | h59b151f_1 14.0 MB conda-forge
ipydatawidgets-4.2.0 | pyhd3deb0d_0 171 KB conda-forge
ipyleaflet-0.13.4 | pyhd3deb0d_0 4.3 MB conda-forge
ipympl-0.5.8 | pyh9f0ad1d_0 506 KB conda-forge
ipyvolume-0.6.0a6 | pyh9f0ad1d_0 5.1 MB conda-forge
ipyvue-1.5.0 | pyhd3deb0d_0 2.2 MB conda-forge
ipyvuetify-1.6.2 | pyh44b312d_0 7.7 MB conda-forge
ipywebrtc-0.5.0 | py38h32f6830_1 877 KB conda-forge
jmespath-0.10.0 | pyh9f0ad1d_0 21 KB conda-forge
Note: you may need to restart the kernel to use updated packages.
libprotobuf-3.13.0.1 | h200bbdf_0 2.3 MB conda-forge
libthrift-0.13.0 | hdfef310_6 1.6 MB conda-forge
libutf8proc-2.6.1 | hcb41399_0 98 KB conda-forge
openssl-1.1.1h | he774522_0 5.8 MB conda-forge
parquet-cpp-1.5.1 | 2 3 KB conda-forge
pcre-8.44 | ha925a31_0 498 KB conda-forge
progressbar2-3.53.1 | pyh9f0ad1d_0 25 KB conda-forge
pyarrow-2.0.0 |py38ha37a76c_6_cpu 2.0 MB conda-forge
python-utils-2.5.5 | pyh44b312d_0 15 KB conda-forge
python_abi-3.8 | 1_cp38 4 KB conda-forge
pythreejs-2.3.0 | pyhd8ed1ab_0 2.6 MB conda-forge
re2-2020.11.01 | h0e60522_0 468 KB conda-forge
s3fs-0.2.2 | py_0 20 KB conda-forge
s3transfer-0.3.4 | pyhd8ed1ab_0 51 KB conda-forge
shapely-1.7.1 | py38h2426642_4 420 KB conda-forge
snappy-1.1.8 | ha925a31_3 50 KB conda-forge
tabulate-0.8.9 | pyhd8ed1ab_0 26 KB conda-forge
traittypes-0.2.1 | pyh9f0ad1d_2 10 KB conda-forge
vaex-3.0.0 | pyh9f0ad1d_0 9 KB conda-forge
vaex-arrow-0.5.1 | pyh9f0ad1d_0 10 KB conda-forge
vaex-astro-0.7.0 | pyh9f0ad1d_0 13 KB conda-forge
vaex-core-2.0.3 | py38h4c96930_1 1.5 MB conda-forge
vaex-hdf5-0.6.0 | pyh9f0ad1d_0 13 KB conda-forge
vaex-jupyter-0.5.2 | pyh9f0ad1d_0 36 KB conda-forge
vaex-ml-0.9.0 | pyh9f0ad1d_0 76 KB conda-forge
vaex-server-0.3.1 | pyh9f0ad1d_0 14 KB conda-forge
vaex-viz-0.4.0 | pyh9f0ad1d_0 18 KB conda-forge
xarray-0.17.0 | pyhd8ed1ab_0 561 KB conda-forge
------------------------------------------------------------
Total: 82.3 MB
The following NEW packages will be INSTALLED:
abseil-cpp conda-forge/win-64::abseil-cpp-20200225.2-ha925a31_2
aplus conda-forge/noarch::aplus-0.11.0-py_1
arrow-cpp conda-forge/win-64::arrow-cpp-2.0.0-py38h647f3f1_6_cpu
aws-c-common conda-forge/win-64::aws-c-common-0.4.59-h8ffe710_1
aws-c-event-stream conda-forge/win-64::aws-c-event-stream-0.1.6-hb4e73fc_6
aws-checksums conda-forge/win-64::aws-checksums-0.1.10-h6f0a1a5_0
aws-sdk-cpp conda-forge/win-64::aws-sdk-cpp-1.8.70-he2782d2_1
boto3 conda-forge/noarch::boto3-1.17.19-pyhd8ed1ab_0
botocore conda-forge/noarch::botocore-1.20.19-pyhd8ed1ab_0
bqplot conda-forge/noarch::bqplot-0.12.20-pyhd8ed1ab_0
branca conda-forge/noarch::branca-0.4.2-pyhd8ed1ab_0
brotli conda-forge/win-64::brotli-1.0.9-h0e60522_4
c-ares conda-forge/win-64::c-ares-1.17.1-h8ffe710_0
cachetools conda-forge/noarch::cachetools-4.2.1-pyhd8ed1ab_0
geos conda-forge/win-64::geos-3.9.1-h39d44d4_2
gflags conda-forge/win-64::gflags-2.2.2-ha925a31_1004
glog conda-forge/win-64::glog-0.4.0-h0174b99_3
grpc-cpp conda-forge/win-64::grpc-cpp-1.33.2-h59b151f_1
ipydatawidgets conda-forge/noarch::ipydatawidgets-4.2.0-pyhd3deb0d_0
ipyleaflet conda-forge/noarch::ipyleaflet-0.13.4-pyhd3deb0d_0
ipympl conda-forge/noarch::ipympl-0.5.8-pyh9f0ad1d_0
ipyvolume conda-forge/noarch::ipyvolume-0.6.0a6-pyh9f0ad1d_0
ipyvue conda-forge/noarch::ipyvue-1.5.0-pyhd3deb0d_0
ipyvuetify conda-forge/noarch::ipyvuetify-1.6.2-pyh44b312d_0
ipywebrtc conda-forge/win-64::ipywebrtc-0.5.0-py38h32f6830_1
jmespath conda-forge/noarch::jmespath-0.10.0-pyh9f0ad1d_0
libprotobuf conda-forge/win-64::libprotobuf-3.13.0.1-h200bbdf_0
libthrift conda-forge/win-64::libthrift-0.13.0-hdfef310_6
libutf8proc conda-forge/win-64::libutf8proc-2.6.1-hcb41399_0
parquet-cpp conda-forge/noarch::parquet-cpp-1.5.1-2
pcre conda-forge/win-64::pcre-8.44-ha925a31_0
progressbar2 conda-forge/noarch::progressbar2-3.53.1-pyh9f0ad1d_0
pyarrow conda-forge/win-64::pyarrow-2.0.0-py38ha37a76c_6_cpu
python-utils conda-forge/noarch::python-utils-2.5.5-pyh44b312d_0
python_abi conda-forge/win-64::python_abi-3.8-1_cp38
pythreejs conda-forge/noarch::pythreejs-2.3.0-pyhd8ed1ab_0
re2 conda-forge/win-64::re2-2020.11.01-h0e60522_0
s3fs conda-forge/noarch::s3fs-0.2.2-py_0
s3transfer conda-forge/noarch::s3transfer-0.3.4-pyhd8ed1ab_0
shapely conda-forge/win-64::shapely-1.7.1-py38h2426642_4
snappy conda-forge/win-64::snappy-1.1.8-ha925a31_3
tabulate conda-forge/noarch::tabulate-0.8.9-pyhd8ed1ab_0
traittypes conda-forge/noarch::traittypes-0.2.1-pyh9f0ad1d_2
vaex conda-forge/noarch::vaex-3.0.0-pyh9f0ad1d_0
vaex-arrow conda-forge/noarch::vaex-arrow-0.5.1-pyh9f0ad1d_0
vaex-astro conda-forge/noarch::vaex-astro-0.7.0-pyh9f0ad1d_0
vaex-core conda-forge/win-64::vaex-core-2.0.3-py38h4c96930_1
vaex-hdf5 conda-forge/noarch::vaex-hdf5-0.6.0-pyh9f0ad1d_0
vaex-jupyter conda-forge/noarch::vaex-jupyter-0.5.2-pyh9f0ad1d_0
vaex-ml conda-forge/noarch::vaex-ml-0.9.0-pyh9f0ad1d_0
vaex-server conda-forge/noarch::vaex-server-0.3.1-pyh9f0ad1d_0
vaex-viz conda-forge/noarch::vaex-viz-0.4.0-pyh9f0ad1d_0
xarray conda-forge/noarch::xarray-0.17.0-pyhd8ed1ab_0
The following packages will be SUPERSEDED by a higher-priority channel:
conda pkgs/main::conda-4.9.2-py38haa95532_0 --> conda-forge::conda-4.9.2-py38haa244fe_0
openssl pkgs/main --> conda-forge
Downloading and Extracting Packages
vaex-core-2.0.3 | 1.5 MB | | 0%
vaex-core-2.0.3 | 1.5 MB | 1 | 1%
vaex-core-2.0.3 | 1.5 MB | ####9 | 49%
vaex-core-2.0.3 | 1.5 MB | ########## | 100%
vaex-core-2.0.3 | 1.5 MB | ########## | 100%
boto3-1.17.19 | 70 KB | | 0%
boto3-1.17.19 | 70 KB | ########## | 100%
boto3-1.17.19 | 70 KB | ########## | 100%
libprotobuf-3.13.0.1 | 2.3 MB | | 0%
libprotobuf-3.13.0.1 | 2.3 MB | #######3 | 73%
libprotobuf-3.13.0.1 | 2.3 MB | ########## | 100%
libprotobuf-3.13.0.1 | 2.3 MB | ########## | 100%
grpc-cpp-1.33.2 | 14.0 MB | | 0%
grpc-cpp-1.33.2 | 14.0 MB | | 0%
grpc-cpp-1.33.2 | 14.0 MB | 2 | 2%
grpc-cpp-1.33.2 | 14.0 MB | 8 | 9%
grpc-cpp-1.33.2 | 14.0 MB | ##1 | 22%
grpc-cpp-1.33.2 | 14.0 MB | ####1 | 41%
grpc-cpp-1.33.2 | 14.0 MB | ###### | 60%
grpc-cpp-1.33.2 | 14.0 MB | ######## | 80%
grpc-cpp-1.33.2 | 14.0 MB | #########9 | 100%
grpc-cpp-1.33.2 | 14.0 MB | ########## | 100%
pythreejs-2.3.0 | 2.6 MB | | 0%
pythreejs-2.3.0 | 2.6 MB | ###### | 60%
pythreejs-2.3.0 | 2.6 MB | ########## | 100%
pythreejs-2.3.0 | 2.6 MB | ########## | 100%
vaex-hdf5-0.6.0 | 13 KB | | 0%
vaex-hdf5-0.6.0 | 13 KB | ########## | 100%
openssl-1.1.1h | 5.8 MB | | 0%
openssl-1.1.1h | 5.8 MB | 1 | 2%
openssl-1.1.1h | 5.8 MB | ###8 | 38%
openssl-1.1.1h | 5.8 MB | ########8 | 88%
openssl-1.1.1h | 5.8 MB | ########## | 100%
ipyvue-1.5.0 | 2.2 MB | | 0%
ipyvue-1.5.0 | 2.2 MB | #######1 | 72%
ipyvue-1.5.0 | 2.2 MB | ########## | 100%
ipyvue-1.5.0 | 2.2 MB | ########## | 100%
xarray-0.17.0 | 561 KB | | 0%
xarray-0.17.0 | 561 KB | ########2 | 83%
xarray-0.17.0 | 561 KB | ########## | 100%
conda-4.9.2 | 3.1 MB | | 0%
conda-4.9.2 | 3.1 MB | #####3 | 54%
conda-4.9.2 | 3.1 MB | ########## | 100%
conda-4.9.2 | 3.1 MB | ########## | 100%
ipympl-0.5.8 | 506 KB | | 0%
ipympl-0.5.8 | 506 KB | ########## | 100%
ipympl-0.5.8 | 506 KB | ########## | 100%
libutf8proc-2.6.1 | 98 KB | | 0%
libutf8proc-2.6.1 | 98 KB | ########## | 100%
aws-checksums-0.1.10 | 51 KB | | 0%
aws-checksums-0.1.10 | 51 KB | ########## | 100%
ipyvolume-0.6.0a6 | 5.1 MB | | 0%
ipyvolume-0.6.0a6 | 5.1 MB | ##9 | 29%
ipyvolume-0.6.0a6 | 5.1 MB | ########1 | 82%
ipyvolume-0.6.0a6 | 5.1 MB | ########## | 100%
snappy-1.1.8 | 50 KB | | 0%
snappy-1.1.8 | 50 KB | ########## | 100%
parquet-cpp-1.5.1 | 3 KB | | 0%
parquet-cpp-1.5.1 | 3 KB | ########## | 100%
brotli-1.0.9 | 882 KB | | 0%
brotli-1.0.9 | 882 KB | ########## | 100%
brotli-1.0.9 | 882 KB | ########## | 100%
arrow-cpp-2.0.0 | 13.4 MB | | 0%
arrow-cpp-2.0.0 | 13.4 MB | | 0%
arrow-cpp-2.0.0 | 13.4 MB | 8 | 9%
arrow-cpp-2.0.0 | 13.4 MB | #8 | 19%
arrow-cpp-2.0.0 | 13.4 MB | ###4 | 35%
arrow-cpp-2.0.0 | 13.4 MB | ####5 | 45%
arrow-cpp-2.0.0 | 13.4 MB | #####7 | 57%
arrow-cpp-2.0.0 | 13.4 MB | ######7 | 68%
arrow-cpp-2.0.0 | 13.4 MB | #######7 | 78%
arrow-cpp-2.0.0 | 13.4 MB | ######### | 91%
arrow-cpp-2.0.0 | 13.4 MB | ########## | 100%
branca-0.4.2 | 26 KB | | 0%
branca-0.4.2 | 26 KB | ########## | 100%
ipyleaflet-0.13.4 | 4.3 MB | | 0%
ipyleaflet-0.13.4 | 4.3 MB | 5 | 5%
ipyleaflet-0.13.4 | 4.3 MB | ######7 | 68%
ipyleaflet-0.13.4 | 4.3 MB | ########## | 100%
ipyleaflet-0.13.4 | 4.3 MB | ########## | 100%
vaex-astro-0.7.0 | 13 KB | | 0%
vaex-astro-0.7.0 | 13 KB | ########## | 100%
gflags-2.2.2 | 80 KB | | 0%
gflags-2.2.2 | 80 KB | ########## | 100%
libthrift-0.13.0 | 1.6 MB | | 0%
libthrift-0.13.0 | 1.6 MB | #####4 | 55%
libthrift-0.13.0 | 1.6 MB | ########## | 100%
libthrift-0.13.0 | 1.6 MB | ########## | 100%
abseil-cpp-20200225. | 1.9 MB | | 0%
abseil-cpp-20200225. | 1.9 MB | | 1%
abseil-cpp-20200225. | 1.9 MB | ########## | 100%
abseil-cpp-20200225. | 1.9 MB | ########## | 100%
progressbar2-3.53.1 | 25 KB | | 0%
progressbar2-3.53.1 | 25 KB | ########## | 100%
pcre-8.44 | 498 KB | | 0%
pcre-8.44 | 498 KB | ########## | 100%
pcre-8.44 | 498 KB | ########## | 100%
pyarrow-2.0.0 | 2.0 MB | | 0%
pyarrow-2.0.0 | 2.0 MB | | 1%
pyarrow-2.0.0 | 2.0 MB | #########2 | 93%
pyarrow-2.0.0 | 2.0 MB | ########## | 100%
c-ares-1.17.1 | 109 KB | | 0%
c-ares-1.17.1 | 109 KB | ########## | 100%
c-ares-1.17.1 | 109 KB | ########## | 100%
shapely-1.7.1 | 420 KB | | 0%
shapely-1.7.1 | 420 KB | ########## | 100%
shapely-1.7.1 | 420 KB | ########## | 100%
aws-sdk-cpp-1.8.70 | 2.9 MB | | 0%
aws-sdk-cpp-1.8.70 | 2.9 MB | ##6 | 27%
aws-sdk-cpp-1.8.70 | 2.9 MB | ########6 | 87%
aws-sdk-cpp-1.8.70 | 2.9 MB | ########## | 100%
vaex-server-0.3.1 | 14 KB | | 0%
vaex-server-0.3.1 | 14 KB | ########## | 100%
python-utils-2.5.5 | 15 KB | | 0%
python-utils-2.5.5 | 15 KB | ########## | 100%
vaex-viz-0.4.0 | 18 KB | | 0%
vaex-viz-0.4.0 | 18 KB | ########## | 100%
s3fs-0.2.2 | 20 KB | | 0%
s3fs-0.2.2 | 20 KB | ########## | 100%
ipyvuetify-1.6.2 | 7.7 MB | | 0%
ipyvuetify-1.6.2 | 7.7 MB | 4 | 5%
ipyvuetify-1.6.2 | 7.7 MB | #8 | 19%
ipyvuetify-1.6.2 | 7.7 MB | ##3 | 24%
ipyvuetify-1.6.2 | 7.7 MB | ####1 | 41%
ipyvuetify-1.6.2 | 7.7 MB | #####5 | 55%
ipyvuetify-1.6.2 | 7.7 MB | #######7 | 78%
ipyvuetify-1.6.2 | 7.7 MB | #########6 | 96%
ipyvuetify-1.6.2 | 7.7 MB | ########## | 100%
aws-c-common-0.4.59 | 151 KB | | 0%
aws-c-common-0.4.59 | 151 KB | ########## | 100%
aws-c-common-0.4.59 | 151 KB | ########## | 100%
geos-3.9.1 | 1.1 MB | | 0%
geos-3.9.1 | 1.1 MB | ########2 | 82%
geos-3.9.1 | 1.1 MB | ########## | 100%
s3transfer-0.3.4 | 51 KB | | 0%
s3transfer-0.3.4 | 51 KB | ########## | 100%
vaex-jupyter-0.5.2 | 36 KB | | 0%
vaex-jupyter-0.5.2 | 36 KB | ########## | 100%
glog-0.4.0 | 83 KB | | 0%
glog-0.4.0 | 83 KB | ########## | 100%
jmespath-0.10.0 | 21 KB | | 0%
jmespath-0.10.0 | 21 KB | ########## | 100%
vaex-ml-0.9.0 | 76 KB | | 0%
vaex-ml-0.9.0 | 76 KB | ########## | 100%
aws-c-event-stream-0 | 26 KB | | 0%
aws-c-event-stream-0 | 26 KB | ########## | 100%
ipydatawidgets-4.2.0 | 171 KB | | 0%
ipydatawidgets-4.2.0 | 171 KB | ########## | 100%
ipydatawidgets-4.2.0 | 171 KB | ########## | 100%
bqplot-0.12.20 | 1.0 MB | | 0%
bqplot-0.12.20 | 1.0 MB | 1 | 2%
bqplot-0.12.20 | 1.0 MB | ########## | 100%
bqplot-0.12.20 | 1.0 MB | ########## | 100%
tabulate-0.8.9 | 26 KB | | 0%
tabulate-0.8.9 | 26 KB | ########## | 100%
vaex-3.0.0 | 9 KB | | 0%
vaex-3.0.0 | 9 KB | ########## | 100%
aplus-0.11.0 | 6 KB | | 0%
aplus-0.11.0 | 6 KB | ########## | 100%
vaex-arrow-0.5.1 | 10 KB | | 0%
vaex-arrow-0.5.1 | 10 KB | ########## | 100%
botocore-1.20.19 | 4.5 MB | | 0%
botocore-1.20.19 | 4.5 MB | 9 | 9%
botocore-1.20.19 | 4.5 MB | ##7 | 28%
botocore-1.20.19 | 4.5 MB | #####1 | 52%
botocore-1.20.19 | 4.5 MB | #######6 | 77%
botocore-1.20.19 | 4.5 MB | ########## | 100%
botocore-1.20.19 | 4.5 MB | ########## | 100%
python_abi-3.8 | 4 KB | | 0%
python_abi-3.8 | 4 KB | ########## | 100%
cachetools-4.2.1 | 13 KB | | 0%
cachetools-4.2.1 | 13 KB | ########## | 100%
traittypes-0.2.1 | 10 KB | | 0%
traittypes-0.2.1 | 10 KB | ########## | 100%
re2-2020.11.01 | 468 KB | | 0%
re2-2020.11.01 | 468 KB | ########## | 100%
re2-2020.11.01 | 468 KB | ########## | 100%
ipywebrtc-0.5.0 | 877 KB | | 0%
ipywebrtc-0.5.0 | 877 KB | ########7 | 88%
ipywebrtc-0.5.0 | 877 KB | ########## | 100%
Preparing transaction: ...working... done
Verifying transaction: ...working... done
Executing transaction: ...working... done
runfile('C:/Users/sutei/Desktop/CASEY/vaex2.py', wdir='C:/Users/sutei/Desktop/CASEY')
WARNING: This is not valid Python code. If you want to use IPython magics, flexible indentation, and prompt removal, please save this file with the .ipy extension. This will be an error in a future version of Spyder.
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 100000 entries, 0 to 99999
Data columns (total 10 columns):
Column Non-Null Count Dtype
0 c0 100000 non-null int32
1 c1 100000 non-null int32
2 c2 100000 non-null int32
3 c3 100000 non-null int32
4 c4 100000 non-null int32
5 c5 100000 non-null int32
6 c6 100000 non-null int32
7 c7 100000 non-null int32
8 c8 100000 non-null int32
9 c9 100000 non-null int32
dtypes: int32(10)
memory usage: 3.8 MB
Traceback (most recent call last):
File "C:\Users\sutei\Desktop\CASEY\vaex2.py", line 25, in
vaex_df = vaex.from_csv(filepath, convert = True, chunk_size = 5_000_000)
AttributeError: module 'vaex' has no attribute 'from_csv'
Code:
import vaex
import pandas as pd
import numpy as np
n_rows = 100000
n_cols = 10
df = pd.DataFrame(np.random.randint(0, 100, size=(n_rows, n_cols)), columns=['c%d' % i for i in range(n_cols)])
df.info(memory_usage='deep')
creating .csv files
filepath = 'main_dataset.csv'
df.to_csv(filepath, index=False)
create hdf5 files
vaex_df = vaex.from_csv(filepath, convert = True, chunk_size = 5_000_000)
type(vaex_df)