Pytorch Medical Segmentation
Read Chinese Introduction:Here!
Recent Updates
- 2021.1.8 The train and test codes are released.
- 2021.2.6 A bug in dice was fixed with the help of Shanshan Li.
Requirements
- pytorch1.7
- torchio<=0.18.20
- python>=3.6
Notice
- You can modify hparam.py to determine whether 2D or 3D segmentation and whether multicategorization is possible.
- We provide algorithms for almost all 2D and 3D segmentation.
- This repository is compatible with almost all medical data formats(e.g. nii.gz, nii, mhd, nrrd, ...), by modifying fold_arch in hparam.py of the config.
- If you want to use a multi-category program, please modify the following codes yourself. I cannot identify your specific categories.
- https://github.com/MontaEllis/Pytorch-Medical-Segmentation/blob/48edef7751af8551b7432b5491f4cf1964bd0cfc/hparam.py#L6
- https://github.com/MontaEllis/Pytorch-Medical-Segmentation/blob/48edef7751af8551b7432b5491f4cf1964bd0cfc/main.py#L235
- https://github.com/MontaEllis/Pytorch-Medical-Segmentation/blob/48edef7751af8551b7432b5491f4cf1964bd0cfc/main.py#L336
- https://github.com/MontaEllis/Pytorch-Medical-Segmentation/blob/48edef7751af8551b7432b5491f4cf1964bd0cfc/main.py#L496
- https://github.com/MontaEllis/Pytorch-Medical-Segmentation/blob/48edef7751af8551b7432b5491f4cf1964bd0cfc/data_function.py#L69
- https://github.com/MontaEllis/Pytorch-Medical-Segmentation/blob/48edef7751af8551b7432b5491f4cf1964bd0cfc/data_function.py#L167
- Whether in 2D or 3D, this project is processed using patch. Therefore, images do not have to be strictly the same size.
Prepare Your Dataset
Example1
if your source dataset is :
source_dataset
├── source_1.mhd
├── source_1.zraw
├── source_2.mhd
├── source_2.zraw
├── source_3.mhd
├── source_3.zraw
├── source_4.mhd
├── source_4.zraw
└── ...
and your label dataset is :
label_dataset
├── label_1.mhd
├── label_1.zraw
├── label_2.mhd
├── label_2.zraw
├── label_3.mhd
├── label_3.zraw
├── label_4.mhd
├── label_4.zraw
└── ...
then your should modify fold_arch as *.mhd, source_train_dir as source_dataset and label_train_dir as label_dataset in hparam.py
Example2
if your source dataset is :
source_dataset
├── 1
├── source_1.mhd
├── source_1.zraw
├── 2
├── source_2.mhd
├── source_2.zraw
├── 3
├── source_3.mhd
├── source_3.zraw
├── 4
├── source_4.mhd
├── source_4.zraw
└── ...
and your label dataset is :
label_dataset
├── 1
├── label_1.mhd
├── label_1.zraw
├── 2
├── label_2.mhd
├── label_2.zraw
├── 3
├── label_3.mhd
├── label_3.zraw
├── 4
├── label_4.mhd
├── label_4.zraw
└── ...
then your should modify fold_arch as */*.mhd, source_train_dir as source_dataset and label_train_dir as label_dataset in hparam.py
Training
- without pretrained-model
set hparam.train_or_test to 'train'
python main.py
- with pretrained-model
set hparam.train_or_test to 'train'
python main.py -k True
Inference
- testing
set hparam.train_or_test to 'test'
python main.py
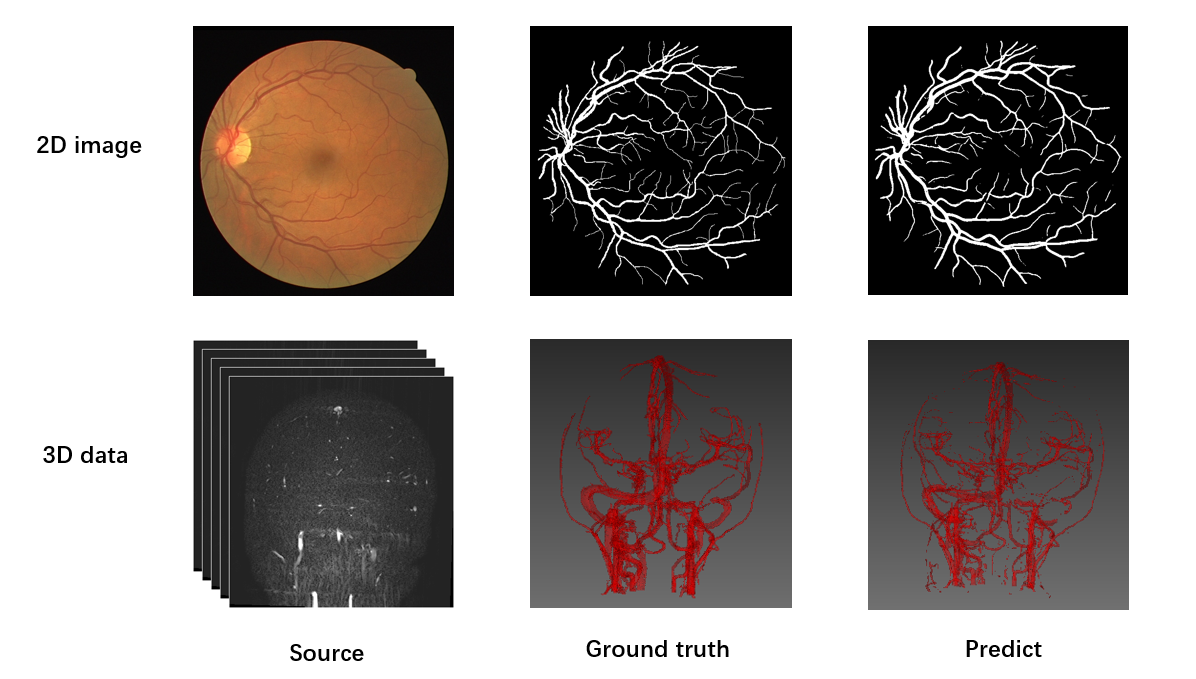
Examples
Done
- 2D
- unet
- unet++
- miniseg
- segnet
- pspnet
- highresnet(copy from https://github.com/fepegar/highresnet, Thank you to fepegar for your generosity!)
- deeplab
- fcn
- 3D
- unet3d
- densevoxelnet3d
- fcn3d
- vnet3d
- highresnert(copy from https://github.com/fepegar/highresnet, Thank you to fepegar for your generosity!)
- densenet3d
TODO
- metrics.py to evaluate your results
- dataset
- benchmark
- nnunet
By The Way
This project is not perfect and there are still many problems. If you are using this project and would like to give the author some feedbacks, you can send Kangneng Zhou an email, his wechat number is: ellisgege666
Acknowledgements
This repository is an unoffical PyTorch implementation of Medical segmentation in 3D and 2D and highly based on MedicalZooPytorch and torchio.Thank you for the above repo. Thank you to Cheng Chen, Daiheng Gao, Jie Zhang, Xing Tao, Weili Jiang and Shanshan Li for all the help I received.
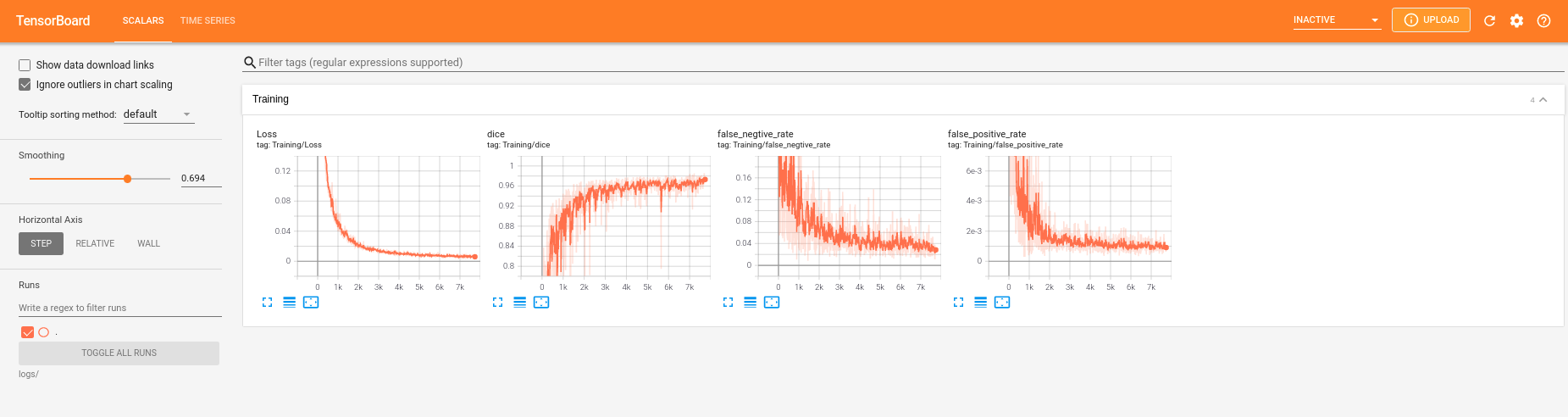
 但是我训练到后面,loss值是在下降,但是dice值就一直是0了,false_positive_rate以及false_negative_rate也很不正常,如下所示:
但是我训练到后面,loss值是在下降,但是dice值就一直是0了,false_positive_rate以及false_negative_rate也很不正常,如下所示:
 想请问原因可以能为什么呢?是否和epoch数有关?因为我看您的原代码是500000,我设置100已经需要跑一段时间了。
其他的参数是否有要根据数据的三维尺寸调整的呢?
感谢回复!
想请问原因可以能为什么呢?是否和epoch数有关?因为我看您的原代码是500000,我设置100已经需要跑一段时间了。
其他的参数是否有要根据数据的三维尺寸调整的呢?
感谢回复!

