OctaDist
Octahedral distortion calculator: A tool for calculating distortion parameters in coordination complexes. https://octadist.github.io/
Register for OctaDist
To get notified when we release new version of OctaDist, please register at https://cutt.ly/regis-octadist.
OctaDist Forum
The users can post questions in our Google Groups: OctaDist Forum
Standard abilities
OctaDist is computer software for inorganic chemistry and crystallography program. OctaDist can be used for studying the structural distortion in coordination complexes. With the abilities of OctaDist, you can:
- analyze the structure and conformation of coordination complexes.
- compute the octahedral distortion parameters.
- explore tilting distortion in perovskite and metal-organic framework.
- display 3D molecule for graphical analysis.
- implement OctaDist's module into your or other program.
- access the program core directly via an interactive scripting language.
Development and Release
OctaDist is written entirely in Python 3 binding to Tkinter GUI toolkit. It is cross-platform program which can work on multiple operating systems. The stable version and development build of OctaDist are released at here. A standalone executable for graphical user interface (GUI) and source code for command line interface (CLI) are available for as follows:
| Platform | Description | Status |
|---|---|---|
| Windows |  |
 |
| Linux |  |
 |
| macOS |  |
 |
| PyPI library |  |
 |
| Anaconda cloud |  |
|
| Nightly build | Development build |  |
Branch:
Git Clone
git clone https://github.com/OctaDist/OctaDist.git
git checkout nightly-build
git pull origin nightly-build
Documents
User manual : https://octadist.github.io/manual.html.
Reference manual :
| Version | Status | Docs |
|---|---|---|
| Stable |  |
HTML / PDF / Epub |
| Dev Build |  |
HTML / PDF / Epub |
Download and Install
For Windows users, we strongly suggest a standalone executable:
Click Here to Download OctaDist-3.0.0-Win-x86-64.exe
For Linux or macOS users and already have Python 3 installed on the system, the easiest way to install OctaDist is to use pip.
pip install octadist
or use conda for those who have Anaconda:
conda install -c rangsiman octadist
Starting OctaDist
The following commands can be used to start OctaDist in different ways:
Graphical User Interface (GUI)
To start GUI program:
octadist
Screenshots of program:
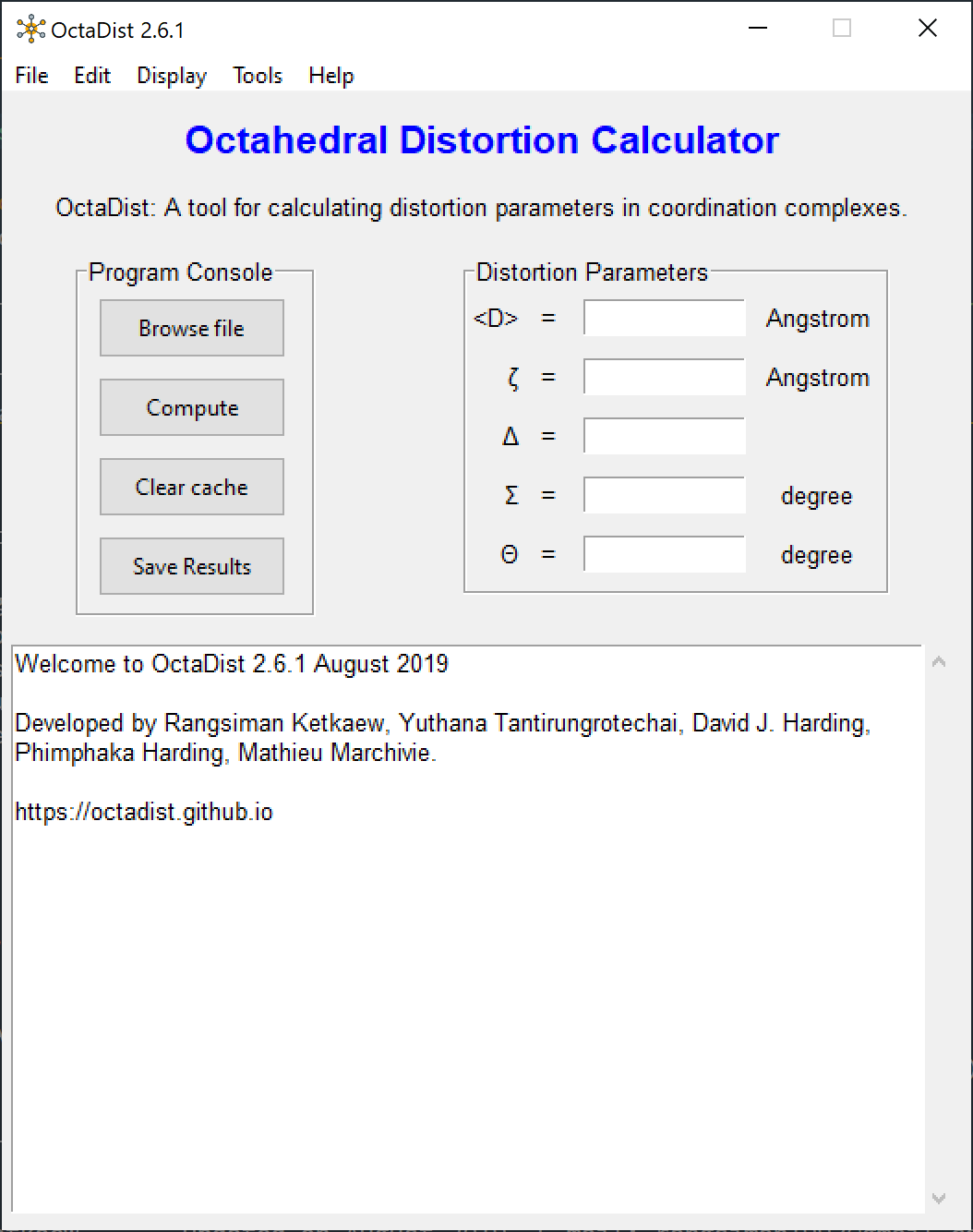 |
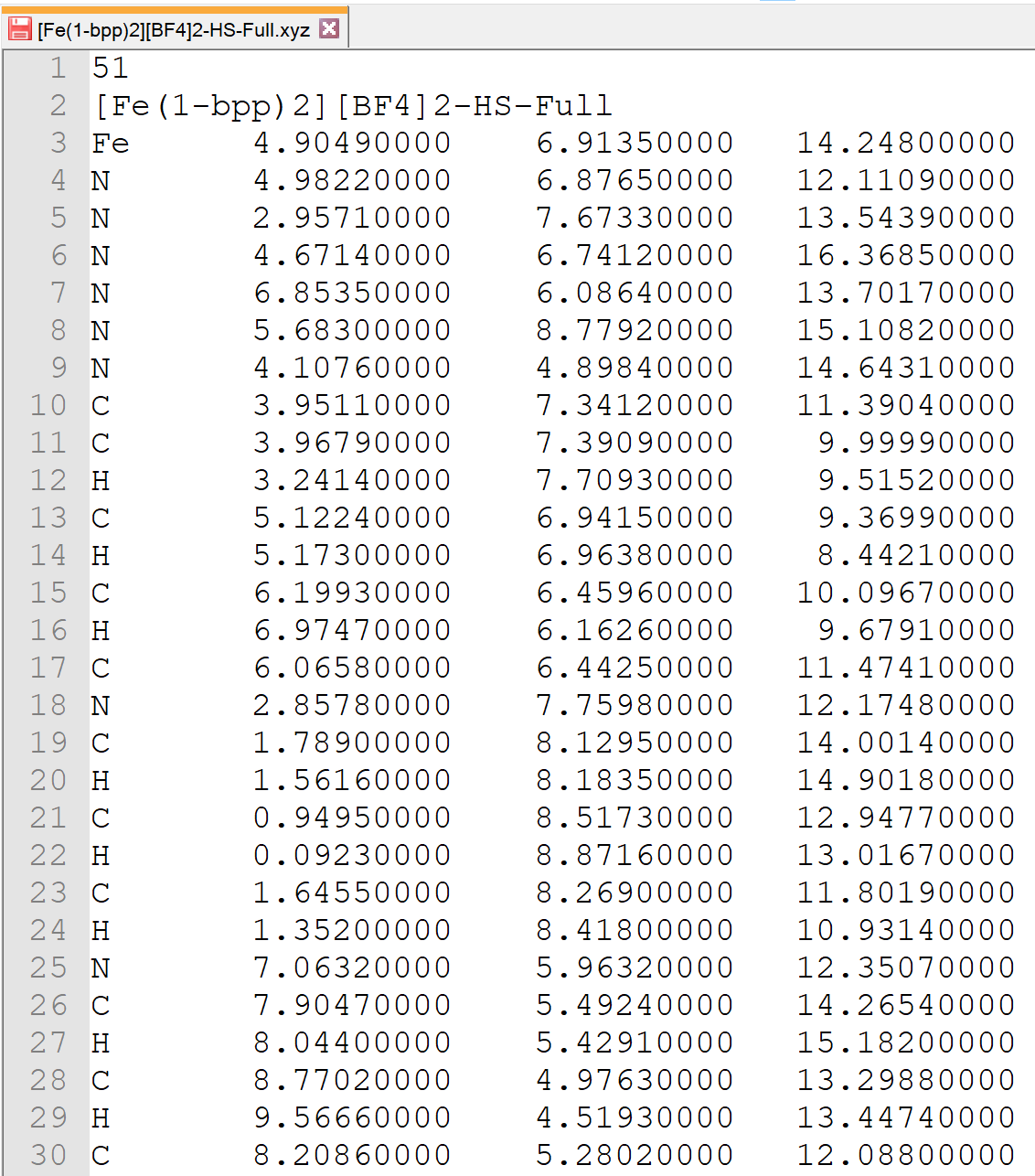 |
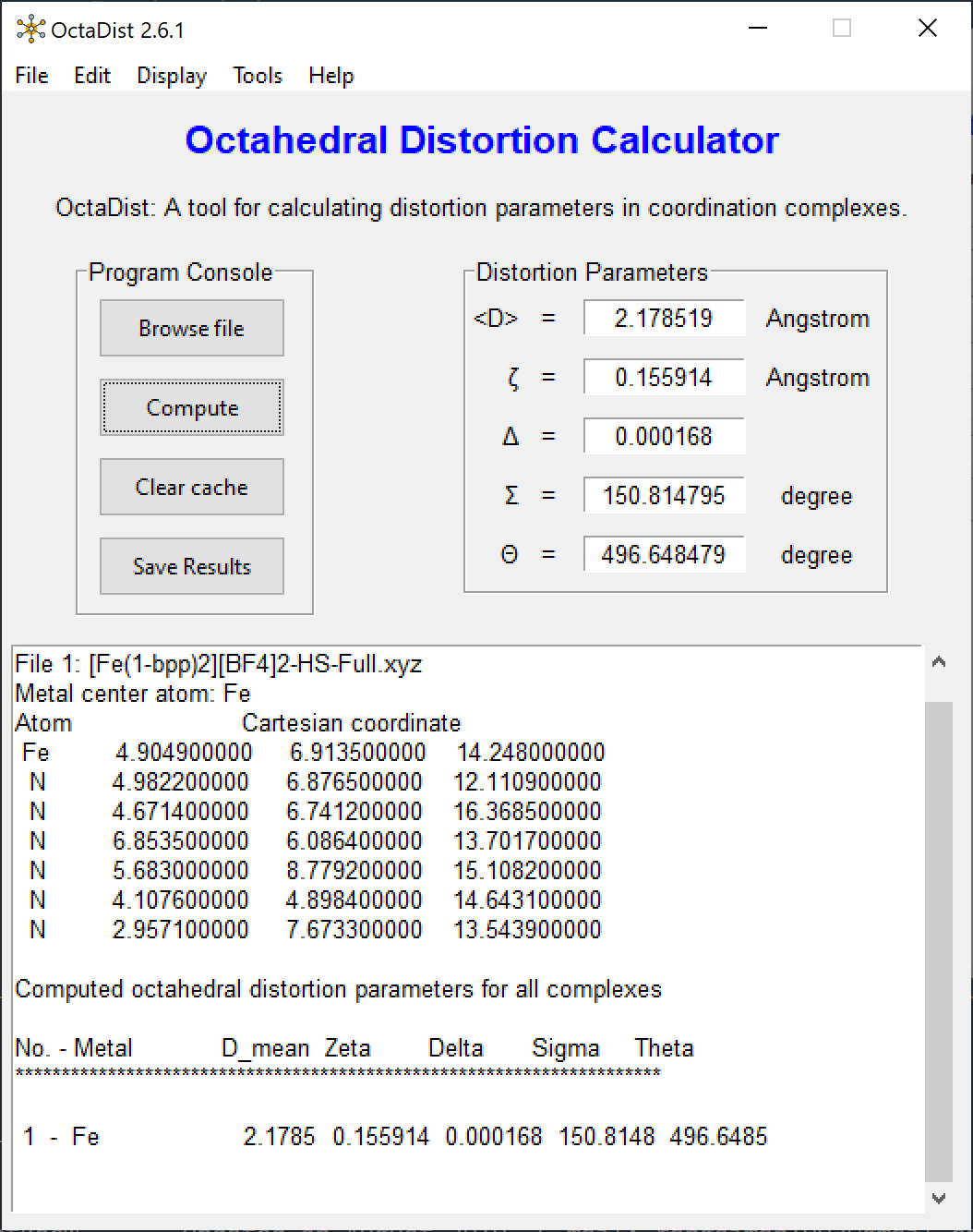 |
|---|---|---|
| OctaDist GUI | XYZ coordinates | Computed distortion parameters |
Command Line Interface (CLI)
To start program command line:
octadist_cli
To calculate distortion parameters:
octadist_cli --inp EXAMPLE_INPUT.xyz
To calculate distortion parameters and show formatted output:
octadist_cli --inp EXAMPLE_INPUT.xyz --out
Supporting input format
Running the tests
Example 1: OctaDist as a package
import octadist as oc
# Prepare list of atomic coordinates of octahedral structure:
atom = ['Fe', 'O', 'O', 'N', 'N', 'N', 'N']
coord = [[2.298354000, 5.161785000, 7.971898000], # <- Metal atom
[1.885657000, 4.804777000, 6.183726000],
[1.747515000, 6.960963000, 7.932784000],
[4.094380000, 5.807257000, 7.588689000],
[0.539005000, 4.482809000, 8.460004000],
[2.812425000, 3.266553000, 8.131637000],
[2.886404000, 5.392925000, 9.848966000]]
dist = oc.CalcDistortion(coord)
zeta = dist.zeta # 0.228072561
delta = dist.delta # 0.000476251
sigma = dist.sigma # 47.92652837
theta = dist.theta # 122.6889727
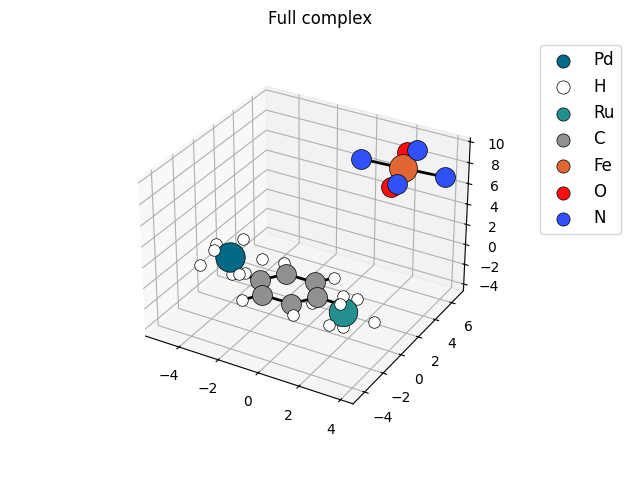
Example 2: Display 3D structure of molecule
import os
import octadist as oc
dir_path = os.path.dirname(os.path.realpath(__file__))
input_folder = os.path.join(dir_path, "../example-input/")
file = input_folder + "Multiple-metals.xyz"
atom_full, coord_full = oc.io.extract_coord(file)
my_plot = oc.draw.DrawComplex_Matplotlib(atom=atom_full, coord=coord_full)
my_plot.add_atom()
my_plot.add_bond()
my_plot.add_legend()
my_plot.save_img()
my_plot.show_plot()
# Figure will be saved as Complex_saved_by_OctaDist.png by default.
Other example scripts and octahedral complexes are available at example-py and example-input, respectively.
Citation
Please cite this project when you use OctaDist for scientific publication.
Ketkaew, R.; Tantirungrotechai, Y.; Harding, P.; Chastanet, G.; Guionneau, P.; Marchivie, M.; Harding, D. J.
OctaDist: A Tool for Calculating Distortion Parameters in Spin Crossover and Coordination Complexes.
Dalton Trans., 2021,50, 1086-1096. https://doi.org/10.1039/D0DT03988H
BibTeX
@article{Ketkaew2021,
doi = {10.1039/d0dt03988h},
url = {https://doi.org/10.1039/d0dt03988h},
year = {2021},
publisher = {Royal Society of Chemistry ({RSC})},
volume = {50},
number = {3},
pages = {1086--1096},
author = {Rangsiman Ketkaew and Yuthana Tantirungrotechai and Phimphaka Harding and Guillaume Chastanet and Philippe Guionneau and Mathieu Marchivie and David J. Harding},
title = {OctaDist: a tool for calculating distortion parameters in spin crossover and coordination complexes},
journal = {Dalton Transactions}
}
Bug report
If you found issues in OctaDist, please report it to us at here.
Project team
- Rangsiman Ketkaew (Thammasat University, Thailand)
- E-mail: [email protected]
- Yuthana Tantirungrotechai (Thammasat University, Thailand)
- E-mail: [email protected]
- Phimphaka Harding (Walailak University, Thailand)
- E-mail: [email protected]
- Guillaume Chastanet (University of Bordeaux, France)
- E-mail: [email protected]
- Philippe Guionneau (University of Bordeaux, France)
- E-mail: [email protected]
- Mathieu Marchivie (University of Bordeaux, France)
- E-mail: [email protected]
- David J. Harding (Walailak University, Thailand)
- E-mail: [email protected]