Ensemble Knowledge Guided Sub-network Search and Fine-tuning for Filter Pruning
This repository is official Tensorflow implementation of paper:
Ensemble Knowledge Guided Sub-network Search and Fine-tuning for Filter Pruning [paper link]
and Tensorflow 2 example code for
"Custom layers", "Custom training loop", "XLA (JIT)-compiling", "Distributed learing", and "Gradients accumulator".
Paper abstract
Conventional NAS-based pruning algorithms aim to find the sub-network with the best validation performance. However, validation performance does not successfully represent test performance, i.e., potential performance. Also, although fine-tuning the pruned network to restore the performance drop is an inevitable process, few studies have handled this issue. This paper proposes a novel sub-network search and fine-tuning method, i.e., Ensemble Knowledge Guidance (EKG). First, we experimentally prove that the fluctuation of the loss landscape is an effective metric to evaluate the potential performance. In order to search a sub-network with the smoothest loss landscape at a low cost, we propose a pseudo-supernet built by an ensemble sub-network knowledge distillation. Next, we propose a novel fine-tuning that re-uses the information of the search phase. We store the interim sub-networks, that is, the by-products of the search phase, and transfer their knowledge into the pruned network. Note that EKG is easy to be plugged-in and computationally efficient. For example, in the case of ResNet-50, about 45% of FLOPS is removed without any performance drop in only 315 GPU hours.

Conceptual visualization of the goal of the proposed method.
Contribution points and key features
- As a new tool to measure the potential performance of sub-network in NAS-based pruning, the smoothness of the loss landscape is presented. Also, the experimental evidence that the loss landscape fluctuation has a higher correlation with the test performance than the validation performance is provided.
- The pseudo-supernet based on an ensemble sub-network knowledge distillation is proposed to find a sub-network of smoother loss landscape without increasing complexity. It helps NAS-based pruning to prune all pre-trained networks, and also allows to find optimal sub-network(s) more accurately.
- To our knowledge, this paper provides the world-first approach to store the information of the search phase in a memory bank and to reuse it in the fine-tuning phase of the pruned network. The proposed memory bank contributes to greatly improving the performance of the pruned network.
- Supernet-based filter pruning code based on Tensorflow2
- Custom layers, e.g., concolution, depthwise convolution, batch normalization (see
nets/tcl.py) - Custom training loop with XLA (JIT) compiling (see
op_utils.py) - distributed learning (see
op_utils.pyanddataloader) - and gradients accumulator (see
op_utils.pyandutils/accumulator)
Requirement
- Tensorflow >= 2.7 (I have tested on 2.7-2.8)
- Pickle
- tqdm
How to run
- Move to the codebase.
- Train and evaluate our model by the below command.
# ResNet-56 on CIFAR10
python train_cifar.py --gpu_id 0 --arch ResNet-56 --dataset CIFAR10 --search_target_rate 0.45 --train_path ../test
python test.py --gpu_id 0 --arch ResNet-56 --dataset CIFAR10 --trained_param ../test/trained_param.pkl
Experimental results

(Left) Potential performance vs. validation loss (right) Potential performance vs. condition number. 50 sub-networks of ResNet-56 trained on CIFAR10 were used for this experiment. accurately.
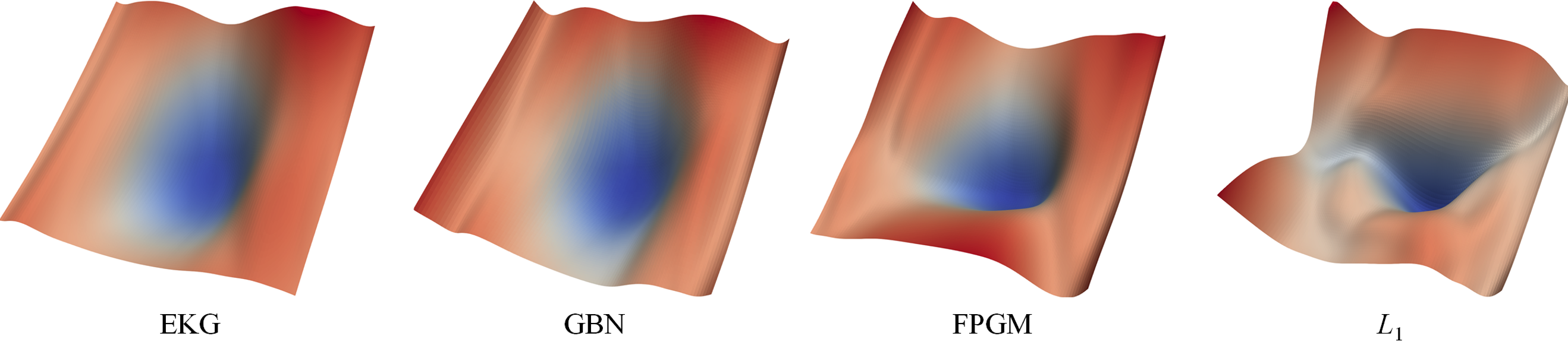
Visualization of loss landscapes of sub-networks searched by various filter importance scoring algorithms.
Comparison with various pruning techniques for ResNet family trained on ImageNet.


Performance analysis in case of ResNet-50 trained on ImageNet-2012. The left plot is the FLOPs reduction rate-Top-1 accuracy, and the right plot is the GPU hours-Top-1 accuracy.
Reference
@article{lee2022ensemble,
title = {Ensemble Knowledge Guided Sub-network Search and Fine-tuning for Filter Pruning},
author = {Seunghyun Lee, Byung Cheol Song},
year = 2022,
journal = {arXiv preprint arXiv:2203.02651}
}