PyNNDescent
PyNNDescent is a Python nearest neighbor descent for approximate nearest neighbors. It provides a python implementation of Nearest Neighbor Descent for k-neighbor-graph construction and approximate nearest neighbor search, as per the paper:
Dong, Wei, Charikar Moses, and Kai Li. "Efficient k-nearest neighbor graph construction for generic similarity measures." Proceedings of the 20th international conference on World wide web. ACM, 2011.
This library supplements that approach with the use of random projection trees for initialisation. This can be particularly useful for the metrics that are amenable to such approaches (euclidean, minkowski, angular, cosine, etc.). Graph diversification is also performed, pruning the longest edges of any triangles in the graph.
Currently this library targets relatively high accuracy (80%-100% accuracy rate) approximate nearest neighbor searches.
Why use PyNNDescent?
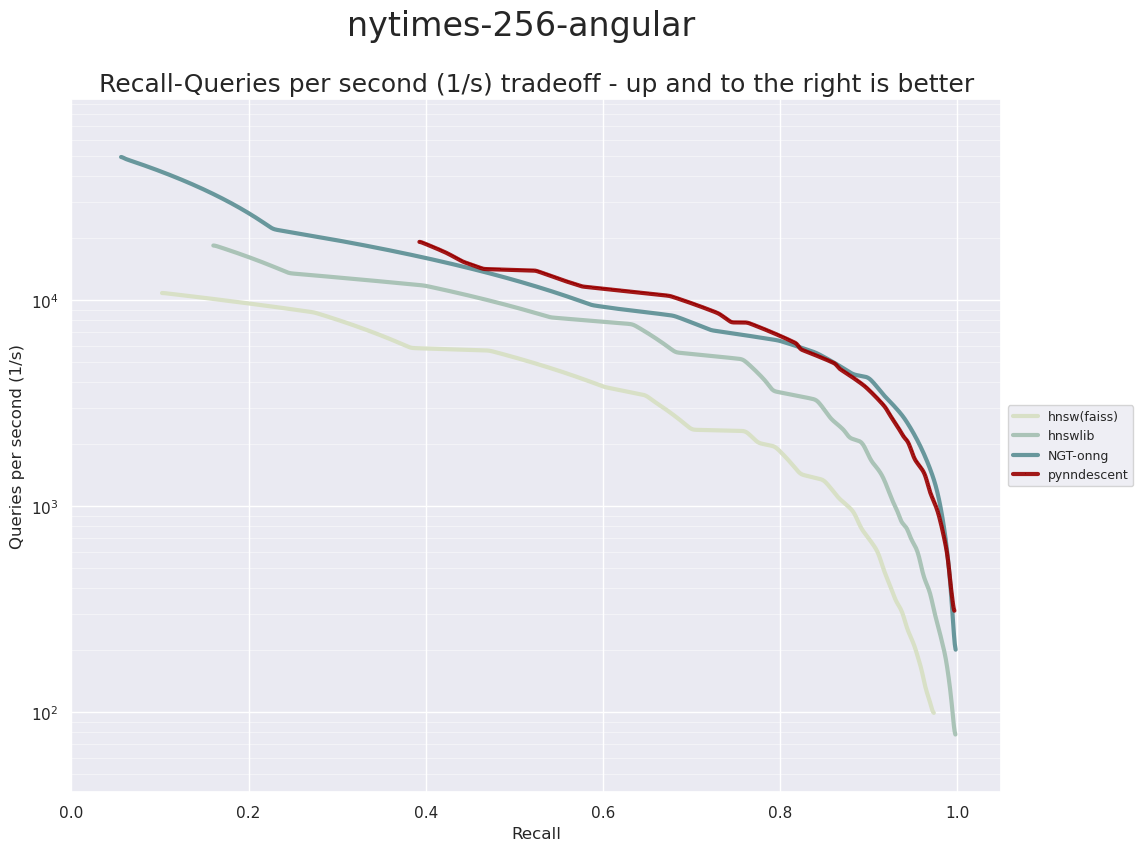
PyNNDescent provides fast approximate nearest neighbor queries. The ann-benchmarks system puts it solidly in the mix of top performing ANN libraries:
SIFT-128 Euclidean
NYTimes-256 Angular
While PyNNDescent is among fastest ANN library, it is also both easy to install (pip and conda installable) with no platform or compilation issues, and is very flexible, supporting a wide variety of distance metrics by default:
Minkowski style metrics
- euclidean
- manhattan
- chebyshev
- minkowski
Miscellaneous spatial metrics
- canberra
- braycurtis
- haversine
Normalized spatial metrics
- mahalanobis
- wminkowski
- seuclidean
Angular and correlation metrics
- cosine
- dot
- correlation
- spearmanr
- tsss
- true_angular
Probability metrics
- hellinger
- wasserstein
Metrics for binary data
- hamming
- jaccard
- dice
- russelrao
- kulsinski
- rogerstanimoto
- sokalmichener
- sokalsneath
- yule
and also custom user defined distance metrics while still retaining performance.
PyNNDescent also integrates well with Scikit-learn, including providing support for the KNeighborTransformer as a drop in replacement for algorithms that make use of nearest neighbor computations.
How to use PyNNDescent
PyNNDescent aims to have a very simple interface. It is similar to (but more limited than) KDTrees and BallTrees in sklearn. In practice there are only two operations -- index construction, and querying an index for nearest neighbors.
To build a new search index on some training data data you can do something like
from pynndescent import NNDescent
index = NNDescent(data)
You can then use the index for searching (and can pickle it to disk if you wish). To search a pynndescent index for the 15 nearest neighbors of a test data set query_data you can do something like
index.query(query_data, k=15)
and that is pretty much all there is to it. You can find more details in the documentation.
Installing
PyNNDescent is designed to be easy to install being a pure python module with relatively light requirements:
- numpy
- scipy
- scikit-learn >= 0.22
- numba >= 0.51
all of which should be pip or conda installable. The easiest way to install should be via conda:
conda install -c conda-forge pynndescent
or via pip:
pip install pynndescent
To manually install this package:
wget https://github.com/lmcinnes/pynndescent/archive/master.zip
unzip master.zip
rm master.zip
cd pynndescent-master
python setup.py install
Help and Support
This project is still young. The documentation is still growing. In the meantime please open an issue and I will try to provide any help and guidance that I can. Please also check the docstrings on the code, which provide some descriptions of the parameters.
License
The pynndescent package is 2-clause BSD licensed. Enjoy.
Contributing
Contributions are more than welcome! There are lots of opportunities for potential projects, so please get in touch if you would like to help out. Everything from code to notebooks to examples and documentation are all equally valuable so please don't feel you can't contribute. To contribute please fork the project make your changes and submit a pull request. We will do our best to work through any issues with you and get your code merged into the main branch.