Describe the bug
Dear colleagues, I am creating a system to classify customers in 2 binary classes and then apply a regression model to one of the classes.
Some of my features are string that I obviously need to encode. In this case with one hot encoding.
To Reproduce
My code is as follows:
from sklearnex import patch_sklearn
patch_sklearn()
from sklearn.pipeline import Pipeline
from sklearn.compose import ColumnTransformer, TransformedTargetRegressor, make_column_selector as selector
from sklearn.preprocessing import StandardScaler, OneHotEncoder, LabelEncoder
from sklearn.impute import SimpleImputer
#from sklearn.model_selection import cross_val_score, cross_validate
from sklearn.multioutput import RegressorChain, MultiOutputRegressor
from sklearn.feature_selection import VarianceThreshold
from sklearn.metrics import mean_absolute_error, make_scorer, mean_tweedie_deviance, auc
from sklearn.model_selection import RandomizedSearchCV, train_test_split, LeaveOneGroupOut, LeavePGroupsOut, cross_validate
from sklearn.metrics import roc_auc_score, plot_roc_curve, roc_curve, confusion_matrix, classification_report, ConfusionMatrixDisplay
from sklearn.ensemble import RandomForestClassifier, HistGradientBoostingRegressor, HistGradientBoostingClassifier
from sklearn import set_config
from mapie.regression import MapieRegressor
data_train, data_test, target_train, target_test = train_test_split(
df.drop(columns=target_reg + target_class + METADATA_COLUMNS),
df[target_reg + target_class],
random_state=42)
categorical_columns_of_interest = ['col1', 'col2', 'col3', 'col4', 'col5', 'col6', 'col7']
numerical_columns = ml_data.drop(columns=target_reg + target_class + METADATA_COLUMNS).select_dtypes(include=np.number).columns
numerical_columns = [x for x in MY_FEATURES if x not in FEATURES_NOT_TO_IMPUTE]
numerical_columns = [x for x in numerical_columns if x not in categorical_columns_of_interest]
categorical_transformer = OneHotEncoder(handle_unknown="ignore")
numeric_transformer = Pipeline(
steps=[
("imputer", SimpleImputer(strategy="mean")),
("scaler", StandardScaler()),
("variance_selector", VarianceThreshold(threshold=0.03))
]
)
preprocessor = ColumnTransformer(
transformers=[
("numeric_only", numeric_transformer, numerical_columns),
("get_dummies", categorical_transformer, categorical_columns_of_interest)])
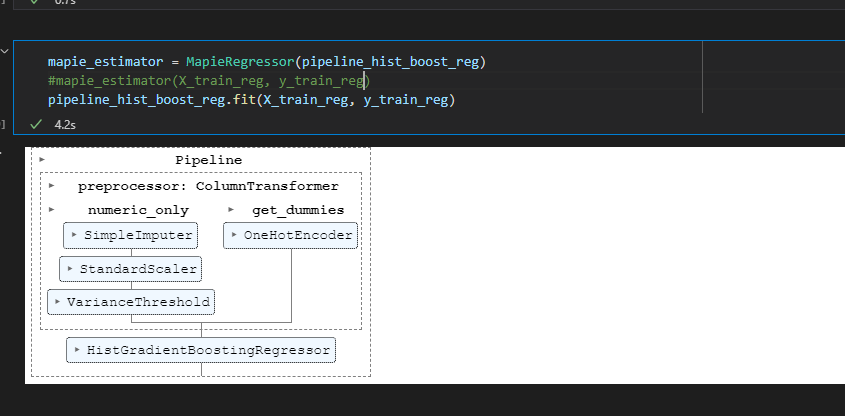
pipeline_hist_boost_reg= Pipeline([('preprocessor', preprocessor),
('estimator', HistGradientBoostingRegressor())])
regressor = TransformedTargetRegressor(pipeline_hist_boost_reg, func=np.log1p, inverse_func=np.expm1)
mapie_estimator = MapieRegressor(pipeline_hist_boost_reg)
mapie_estimator.fit(data_train, target_train)
Expected behavior
After this I will expect that I can run:
y_pred, y_pis = mapie_estimator.predict(data_test)
Screenshots
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
<ipython-input-43-61f70ee71787> in <module>
1 mapie_estimator = MapieRegressor(pipeline_hist_boost_reg)
----> 2 mapie_estimator.fit(X_train_reg, y_train_reg)
/anaconda/envs/azureml_py38/lib/python3.8/site-packages/mapie/regression.py in fit(self, X, y, sample_weight)
457 cv = self._check_cv(self.cv)
458 estimator = self._check_estimator(self.estimator)
--> 459 X, y = check_X_y(
460 X, y, force_all_finite=False, dtype=["float64", "int", "object"]
461 )
/anaconda/envs/azureml_py38/lib/python3.8/site-packages/sklearn/utils/validation.py in check_X_y(X, y, accept_sparse, accept_large_sparse, dtype, order, copy, force_all_finite, ensure_2d, allow_nd, multi_output, ensure_min_samples, ensure_min_features, y_numeric, estimator)
962 raise ValueError("y cannot be None")
963
--> 964 X = check_array(
965 X,
966 accept_sparse=accept_sparse,
/anaconda/envs/azureml_py38/lib/python3.8/site-packages/sklearn/utils/validation.py in check_array(array, accept_sparse, accept_large_sparse, dtype, order, copy, force_all_finite, ensure_2d, allow_nd, ensure_min_samples, ensure_min_features, estimator)
683 if has_pd_integer_array:
684 # If there are any pandas integer extension arrays,
--> 685 array = array.astype(dtype)
686
687 if force_all_finite not in (True, False, "allow-nan"):
/anaconda/envs/azureml_py38/lib/python3.8/site-packages/pandas/core/generic.py in astype(self, dtype, copy, errors)
5804 else:
5805 # else, only a single dtype is given
-> 5806 new_data = self._mgr.astype(dtype=dtype, copy=copy, errors=errors)
5807 return self._constructor(new_data).__finalize__(self, method="astype")
5808
/anaconda/envs/azureml_py38/lib/python3.8/site-packages/pandas/core/internals/managers.py in astype(self, dtype, copy, errors)
412
413 def astype(self: T, dtype, copy: bool = False, errors: str = "raise") -> T:
--> 414 return self.apply("astype", dtype=dtype, copy=copy, errors=errors)
415
416 def convert(
/anaconda/envs/azureml_py38/lib/python3.8/site-packages/pandas/core/internals/managers.py in apply(self, f, align_keys, ignore_failures, **kwargs)
325 applied = b.apply(f, **kwargs)
326 else:
--> 327 applied = getattr(b, f)(**kwargs)
328 except (TypeError, NotImplementedError):
329 if not ignore_failures:
/anaconda/envs/azureml_py38/lib/python3.8/site-packages/pandas/core/internals/blocks.py in astype(self, dtype, copy, errors)
590 values = self.values
591
--> 592 new_values = astype_array_safe(values, dtype, copy=copy, errors=errors)
593
594 new_values = maybe_coerce_values(new_values)
/anaconda/envs/azureml_py38/lib/python3.8/site-packages/pandas/core/dtypes/cast.py in astype_array_safe(values, dtype, copy, errors)
1298
1299 try:
-> 1300 new_values = astype_array(values, dtype, copy=copy)
1301 except (ValueError, TypeError):
1302 # e.g. astype_nansafe can fail on object-dtype of strings
/anaconda/envs/azureml_py38/lib/python3.8/site-packages/pandas/core/dtypes/cast.py in astype_array(values, dtype, copy)
1246
1247 else:
-> 1248 values = astype_nansafe(values, dtype, copy=copy)
1249
1250 # in pandas we don't store numpy str dtypes, so convert to object
/anaconda/envs/azureml_py38/lib/python3.8/site-packages/pandas/core/dtypes/cast.py in astype_nansafe(arr, dtype, copy, skipna)
1083 flags = arr.flags
1084 flat = arr.ravel("K")
-> 1085 result = astype_nansafe(flat, dtype, copy=copy, skipna=skipna)
1086 order: Literal["C", "F"] = "F" if flags.f_contiguous else "C"
1087 # error: Item "ExtensionArray" of "Union[ExtensionArray, ndarray]" has no
/anaconda/envs/azureml_py38/lib/python3.8/site-packages/pandas/core/dtypes/cast.py in astype_nansafe(arr, dtype, copy, skipna)
1190 if copy or is_object_dtype(arr.dtype) or is_object_dtype(dtype):
1191 # Explicit copy, or required since NumPy can't view from / to object.
-> 1192 return arr.astype(dtype, copy=True)
1193
1194 return arr.astype(dtype, copy=copy)
ValueError: could not convert string to float: 'group C'
Being this value part of one of the categorical columns which its being encoded by the preprocessor.
When training the model without mapie everything works correctly:
Desktop (please complete the following information):
import platform
print(platform.machine())
print(platform.version())
print(platform.platform())
print(platform.system())
print(platform.processor())
x86_64
#58~18.04.1-Ubuntu SMP Wed Jul 28 23:14:18 UTC 2021
Linux-5.4.0-1056-azure-x86_64-with-glibc2.10
Linux
x86_64
Scikit learn dependencies:
scikit-learn==1.0.2
scikit-learn-intelex==2021.5.1
imbalance-learn==0.9
mapie==0.3.1
bug









