Overview of OpenPNM
OpenPNM is a comprehensive framework for performing pore network simulations of porous materials.
More Information
For more details about the package can be found in the on-line documentation
Stay Informed
It is surprizingly hard to communicate with our users, since Github doesn't allow sending out email newsletters or announcements. To address this gap, we have created a Substack channel, where you can subscribe to our feed to receive periodic news about important events and updates. Also, follow us on Twitter (@OpenPnm) for periodic announcements about new releases and other important events.
Installation and Requirements
Preferred method
The preferred way of installing OpenPNM is through Anaconda Cloud using:
conda install -c conda-forge openpnm
Alternative method
OpenPNM can also be installed from the Python Package Index using:
pip install openpnm
However, we don't recommend installing using pip since pypardiso, which is a blazing fast direct solver, is not available for Windows users who use Python 3.7+.
For developers
For developers who intend to change the source code or contribute to OpenPNM, the source code can be downloaded from Github and installed by running:
pip install -e 'path/to/downloaded/files'
The advantage to installing from the source code is that you can edit the files and have access to your changes each time you import OpenPNM.
OpenPNM requires the Scipy Stack (Numpy, Scipy, Matplotlib, etc), which is most conveniently obtained by installing the Anaconda Distribution.
Example Usage
The following code block illustrates how to use OpenPNM to perform a mercury intrusion porosimetry simulation:
import openpnm as op
pn = op.network.Cubic(shape=[10, 10, 10], spacing=0.0001)
geo = op.geometry.SpheresAndCylinders(network=pn, pores=pn.Ps, throats=pn.Ts)
Hg = op.phases.Mercury(network=pn)
phys = op.physics.Standard(network=pn, phase=Hg, geometry=geo)
mip = op.algorithms.Porosimetry(network=pn, phase=Hg)
mip.set_inlets(pores=pn.pores(['left', 'right', 'top', 'bottom']))
mip.run()
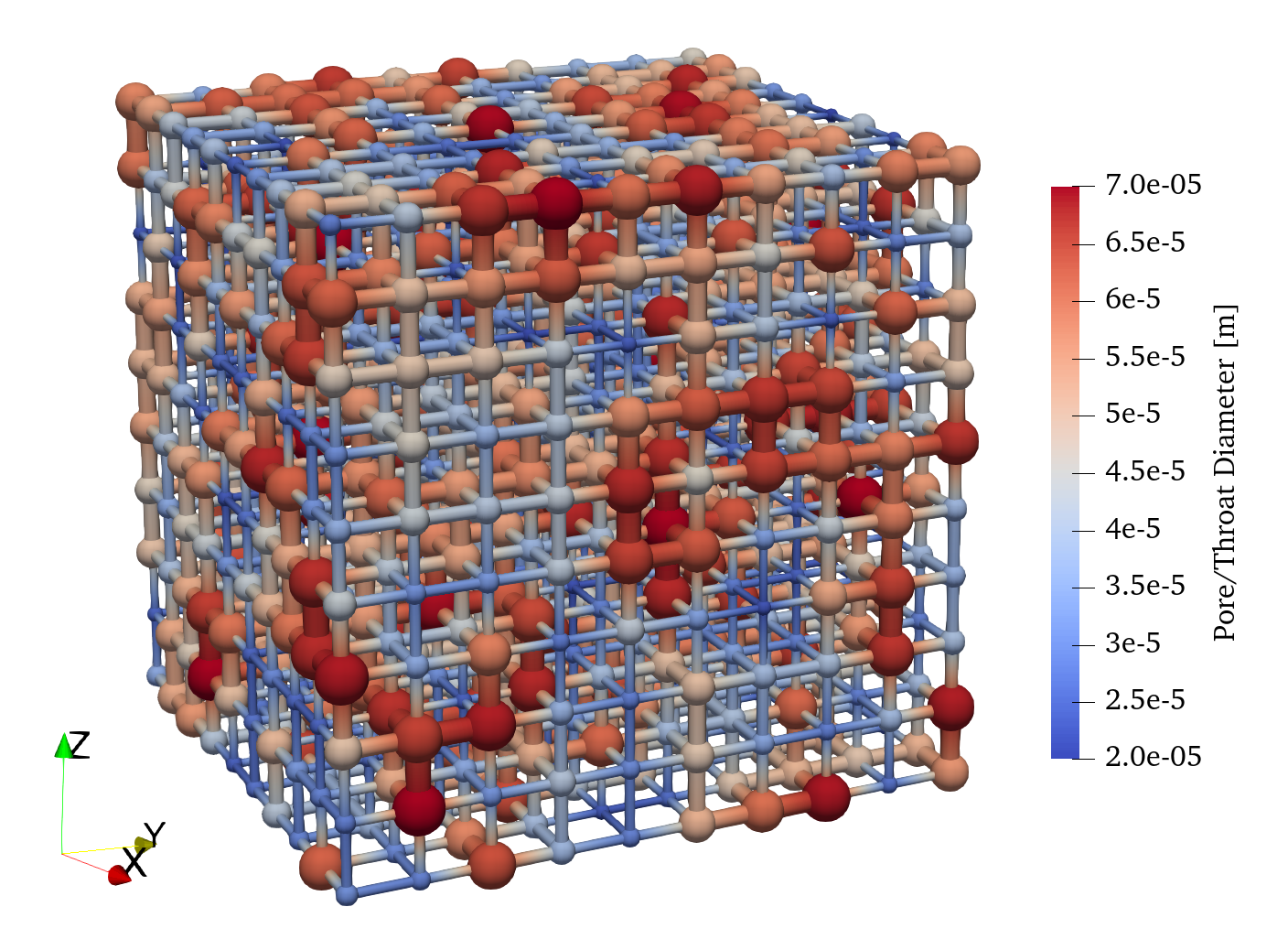
The network can be visualized in ParaView giving the following:
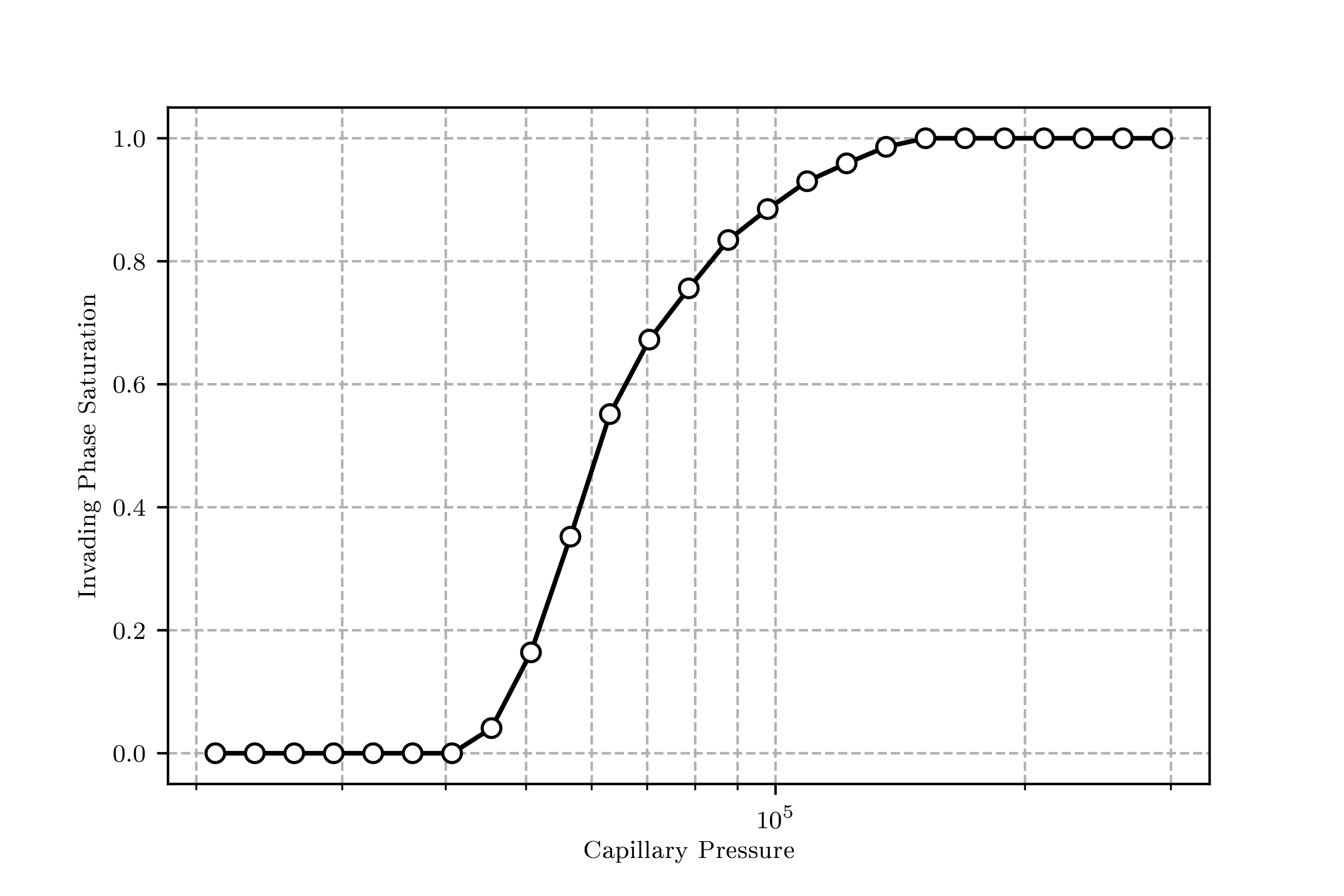
The drainage curve can be visualized with mip.plot_intrusion_curve() giving something like this:
A collection of examples is available in the examples folder of this repository: Examples
Asking Questions and Getting Help
Github now has a Discussions function, which works similarly to stack overflow. Please post your question in the Q&A category so devs or users can provide answers, vote on accepted answers, improve on each other's answers, and generally discuss things. Most importantly, all answers are searchable so eventually, once enough questions have been posted and answered, you can find what you're looking for with a simple search.
Contact
OpenPNM is developed by the Porous Materials Engineering and Analysis Lab (PMEAL), in the Department of Chemical Engineering at the University of Waterloo in Waterloo, Ontario, Canada.
The lead developer for this project is Prof. Jeff Gostick ([email protected]).
Acknowledgements
OpenPNM is grateful to CANARIE for their generous funding over the past few years. We would also like to acknowledge the support of NSERC of Canada for funding many of the student that have contributed to OpenPNM since it's inception in 2011.
Citation
If you use OpenPNM in a publication, please cite the following paper:
Gostick et al. "OpenPNM: a pore network modeling package." Computing in Science & Engineering 18, no. 4 (2016): 60-74. doi:10.1109/MCSE.2016.49
Also, we ask that you "star"