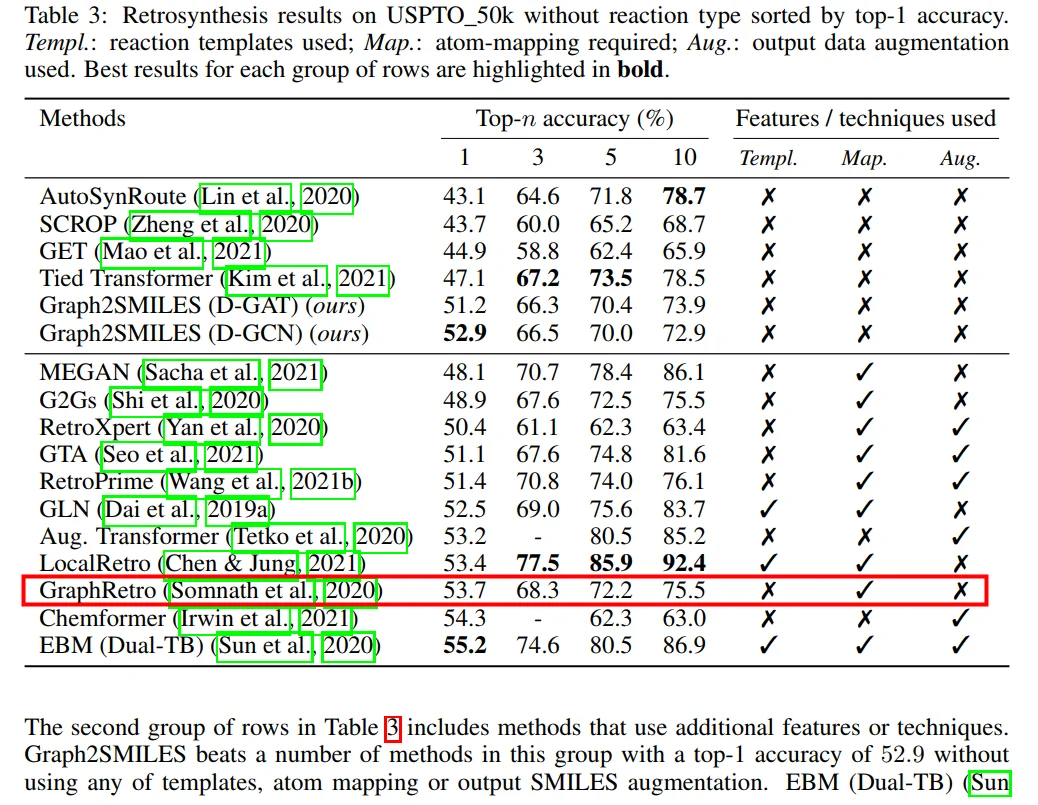
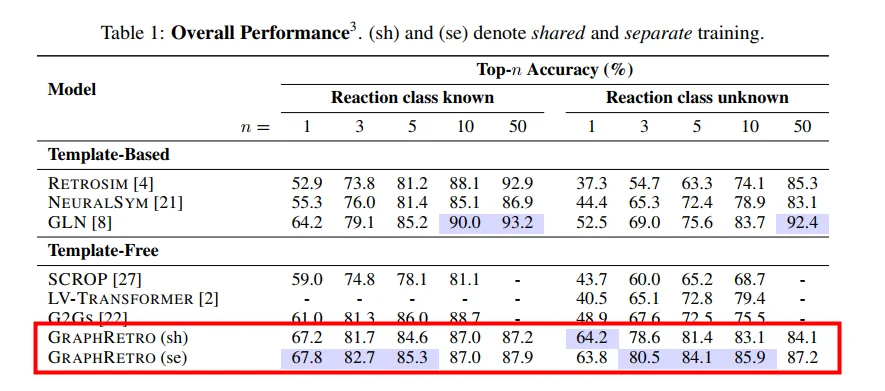
Graph2SMILES
A graph-to-sequence model for one-step retrosynthesis and reaction outcome prediction.
1. Environmental setup
System requirements
Ubuntu: >= 16.04
conda: >= 4.0
GPU: at least 8GB Memory with CUDA >= 10.1
Note: there is some known compatibility issue with RTX 3090, for which the PyTorch would need to be upgraded to >= 1.8.0. The code has not been heavily tested under 1.8.0, so our best advice is to use some other GPU.
Using conda
Please ensure that conda has been properly initialized, i.e. conda activate is runnable. Then
bash -i scripts/setup.sh
conda activate graph2smiles
2. Data preparation
Download the raw (cleaned and tokenized) data from Google Drive by
python scripts/download_raw_data.py --data_name=USPTO_50k
python scripts/download_raw_data.py --data_name=USPTO_full
python scripts/download_raw_data.py --data_name=USPTO_480k
python scripts/download_raw_data.py --data_name=USPTO_STEREO
It is okay to only download the dataset(s) you want. For each dataset, modify the following environmental variables in scripts/preprocess.sh:
DATASET: one of [USPTO_50k, USPTO_full, USPTO_480k, USPTO_STEREO]
TASK: retrosynthesis for 50k and full, or reaction_prediction for 480k and STEREO
N_WORKERS: number of CPU cores (for parallel preprocessing)
Then run the preprocessing script by
sh scripts/preprocess.sh
3. Model training and validation
Modify the following environmental variables in scripts/train_g2s.sh:
EXP_NO: your own identifier (any string) for logging and tracking
DATASET: one of [USPTO_50k, USPTO_full, USPTO_480k, USPTO_STEREO]
TASK: retrosynthesis for 50k and full, or reaction_prediction for 480k and STEREO
MPN_TYPE: one of [dgcn, dgat]
Then run the training script by
sh scripts/train_g2s.sh
The training process regularly evaluates on the validation sets, both with and without teacher forcing. While this evaluation is done mostly with top-1 accuracy, it is also possible to do holistic evaluation after training finishes to get all the top-n accuracies on the val set. To do that, first modify the following environmental variables in scripts/validate.sh:
EXP_NO: your own identifier (any string) for logging and tracking
DATASET: one of [USPTO_50k, USPTO_full, USPTO_480k, USPTO_STEREO]
CHECKPOINT: the folder containing the checkpoints
FIRST_STEP: the step of the first checkpoints to be evaluated
LAST_STEP: the step of the last checkpoints to be evaluated
Then run the evaluation script by
sh scripts/validate.sh
Note: the evaluation process performs beam search over the whole val sets for all checkpoints. It can take tens of hours.
We provide pretrained model checkpoints for all four datasets with both dgcn and dgat, which can be downloaded from Google Drive with
python scripts/download_checkpoints.py --data_name=$DATASET --mpn_type=$MPN_TYPE
using any combinations of DATASET and MPN_TYPE.
4. Testing
Modify the following environmental variables in scripts/predict.sh:
EXP_NO: your own identifier (any string) for logging and tracking
DATASET: one of [USPTO_50k, USPTO_full, USPTO_480k, USPTO_STEREO]
CHECKPOINT: the path to the checkpoint (which is a .pt file)
Then run the testing script by
sh scripts/predict.sh
which will first run beam search to generate the results for all the test inputs, and then computes the average top-n accuracies.