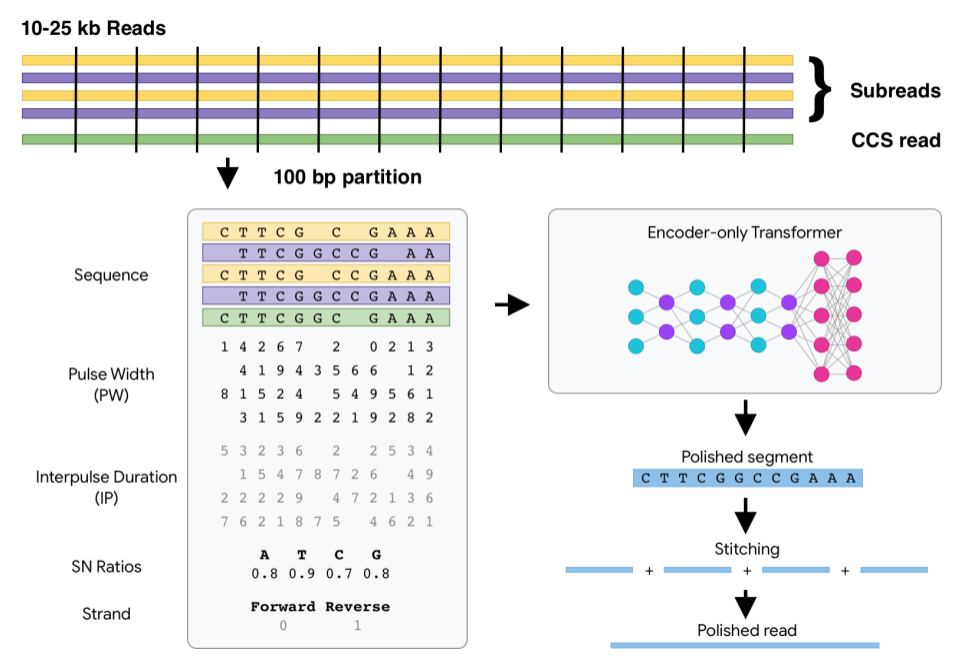
Pango lineage information for German SARS-CoV-2 sequences
This repository contains a join of the metadata and pango lineage tables of all German SARS-CoV-2 sequences published by the Robert-Koch-Institut on Github.
The data here is updated every hour, automatically through a Github action, so whenever new data appears in the RKI repo, you will see it here within at most an hour.
The resulting dataset can be downloaded here, beware it's currently around 50MB in size: https://raw.githubusercontent.com/corneliusroemer/desh-data/main/data/meta_lineages.csv
Omicron share plot
Description of data
Column description:
- IMS_ID: Unique identifier of the sequence
- DATE_DRAW: Date the sample was taken from the patient
- SEQ_REASON: Reason for sequencing, one of:
- X: Unknown
- N: Random sampling
- Y: Targeted sequencing (exact reason unknown)
- A[<reason>]: Targeted sequencing because variant PCR indicated VOC
- PROCESSING_DATE: Date the sample was processed by the RKI and added to Github repo
- SENDING_LAB_PC: Postcode (PLZ) of lab that did the initial PCR
- SEQUENCING_LAB_PC: Postcode (PLZ) of lab that did the sequencing
- lineage: Pango lineage as reported by
pangolin - scorpio_call: Alternative, rough, variant as determined by
scorpio(part ofpangolin), this is less precise but a bit more robust thanpangolin.
Excerpt
Here are the first 10 lines of the dataset.
IMS_ID,DATE_DRAW,SEQ_REASON,PROCESSING_DATE,SENDING_LAB_PC,SEQUENCING_LAB_PC,lineage,scorpio_call
IMS-10294-CVDP-00001,2021-01-14,X,2021-01-25,40225,40225,B.1.1.297,
IMS-10025-CVDP-00001,2021-01-17,N,2021-01-26,10409,10409,B.1.389,
IMS-10025-CVDP-00002,2021-01-17,N,2021-01-26,10409,10409,B.1.258,
IMS-10025-CVDP-00003,2021-01-17,N,2021-01-26,10409,10409,B.1.177.86,
IMS-10025-CVDP-00004,2021-01-17,N,2021-01-26,10409,10409,B.1.389,
IMS-10025-CVDP-00005,2021-01-18,N,2021-01-26,10409,10409,B.1.160,
IMS-10025-CVDP-00006,2021-01-17,N,2021-01-26,10409,10409,B.1.1.297,
IMS-10025-CVDP-00007,2021-01-18,N,2021-01-26,10409,10409,B.1.177.81,
IMS-10025-CVDP-00008,2021-01-18,N,2021-01-26,10409,10409,B.1.177,
IMS-10025-CVDP-00009,2021-01-18,N,2021-01-26,10409,10409,B.1.1.7,Alpha (B.1.1.7-like)
IMS-10025-CVDP-00010,2021-01-17,N,2021-01-26,10409,10409,B.1.1.7,Alpha (B.1.1.7-like)
IMS-10025-CVDP-00011,2021-01-17,N,2021-01-26,10409,10409,B.1.389,
Suggested import into pandas
You can import the data into pandas as follows:
#%%
import pandas as pd
#%%
df = pd.read_csv(
'https://raw.githubusercontent.com/corneliusroemer/desh-data/main/data/meta_lineages.csv',
index_col=0,
parse_dates=[1,3],
infer_datetime_format=True,
cache_dates=True,
dtype = {'SEQ_REASON': 'category',
'SENDING_LAB_PC': 'category',
'SEQUENCING_LAB_PC': 'category',
'lineage': 'category',
'scorpio_call': 'category'
}
)
#%%
df.rename(columns={
'DATE_DRAW': 'date',
'PROCESSING_DATE': 'processing_date',
'SEQ_REASON': 'reason',
'SENDING_LAB_PC': 'sending_pc',
'SEQUENCING_LAB_PC': 'sequencing_pc',
'lineage': 'lineage',
'scorpio_call': 'scorpio'
},
inplace=True
)
df
License
The underlying files that I use as input are licensed by RKI under CC-BY 4.0, see more details here: https://github.com/robert-koch-institut/SARS-CoV-2-Sequenzdaten_aus_Deutschland#lizenz.
The software here is licensed under the "Unlicense". You can do with it whatever you want.
For the data, just cite the original source, no need to cite this repo since it's just a trivial join.