UMAP
Uniform Manifold Approximation and Projection (UMAP) is a dimension reduction technique that can be used for visualisation similarly to t-SNE, but also for general non-linear dimension reduction. The algorithm is founded on three assumptions about the data:
- The data is uniformly distributed on a Riemannian manifold;
- The Riemannian metric is locally constant (or can be approximated as such);
- The manifold is locally connected.
From these assumptions it is possible to model the manifold with a fuzzy topological structure. The embedding is found by searching for a low dimensional projection of the data that has the closest possible equivalent fuzzy topological structure.
The details for the underlying mathematics can be found in our paper on ArXiv:
McInnes, L, Healy, J, UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction, ArXiv e-prints 1802.03426, 2018
The important thing is that you don't need to worry about that—you can use UMAP right now for dimension reduction and visualisation as easily as a drop in replacement for scikit-learn's t-SNE.
Documentation is available via Read the Docs.
New: this package now also provides support for densMAP. The densMAP algorithm augments UMAP to preserve local density information in addition to the topological structure of the data. Details of this method are described in the following paper:
Narayan, A, Berger, B, Cho, H, Density-Preserving Data Visualization Unveils Dynamic Patterns of Single-Cell Transcriptomic Variability, bioRxiv, 2020
Installing
UMAP depends upon scikit-learn, and thus scikit-learn's dependencies such as numpy and scipy. UMAP adds a requirement for numba for performance reasons. The original version used Cython, but the improved code clarity, simplicity and performance of Numba made the transition necessary.
Requirements:
- Python 3.6 or greater
- numpy
- scipy
- scikit-learn
- numba
Recommended packages:
- pynndescent
-
- For plotting
-
- matplotlib
- datashader
- holoviews
-
- for Parametric UMAP
-
- tensorflow > 2.0.0
Installing pynndescent can significantly increase performance, and in later versions it will become a hard dependency.
Install Options
Conda install, via the excellent work of the conda-forge team:
conda install -c conda-forge umap-learn
The conda-forge packages are available for Linux, OS X, and Windows 64 bit.
PyPI install, presuming you have numba and sklearn and all its requirements (numpy and scipy) installed:
pip install umap-learn
If you wish to use the plotting functionality you can use
pip install umap-learn[plot]
to install all the plotting dependencies.
If you wish to use Parametric UMAP, you need to install Tensorflow, which can be installed either using the instructions at https://www.tensorflow.org/install (reccomended) or using
pip install umap-learn[parametric_umap]
for a CPU-only version of Tensorflow.
If pip is having difficulties pulling the dependencies then we'd suggest installing the dependencies manually using anaconda followed by pulling umap from pip:
conda install numpy scipy
conda install scikit-learn
conda install numba
pip install umap-learn
For a manual install get this package:
wget https://github.com/lmcinnes/umap/archive/master.zip
unzip master.zip
rm master.zip
cd umap-master
Install the requirements
sudo pip install -r requirements.txt
or
conda install scikit-learn numba
Install the package
python setup.py install
How to use UMAP
The umap package inherits from sklearn classes, and thus drops in neatly next to other sklearn transformers with an identical calling API.
import umap
from sklearn.datasets import load_digits
digits = load_digits()
embedding = umap.UMAP().fit_transform(digits.data)
There are a number of parameters that can be set for the UMAP class; the major ones are as follows:
n_neighbors: This determines the number of neighboring points used in local approximations of manifold structure. Larger values will result in more global structure being preserved at the loss of detailed local structure. In general this parameter should often be in the range 5 to 50, with a choice of 10 to 15 being a sensible default.min_dist: This controls how tightly the embedding is allowed compress points together. Larger values ensure embedded points are more evenly distributed, while smaller values allow the algorithm to optimise more accurately with regard to local structure. Sensible values are in the range 0.001 to 0.5, with 0.1 being a reasonable default.metric: This determines the choice of metric used to measure distance in the input space. A wide variety of metrics are already coded, and a user defined function can be passed as long as it has been JITd by numba.
An example of making use of these options:
import umap
from sklearn.datasets import load_digits
digits = load_digits()
embedding = umap.UMAP(n_neighbors=5,
min_dist=0.3,
metric='correlation').fit_transform(digits.data)
UMAP also supports fitting to sparse matrix data. For more details please see the UMAP documentation
Benefits of UMAP
UMAP has a few signficant wins in its current incarnation.
First of all UMAP is fast. It can handle large datasets and high dimensional data without too much difficulty, scaling beyond what most t-SNE packages can manage. This includes very high dimensional sparse datasets. UMAP has successfully been used directly on data with over a million dimensions.
Second, UMAP scales well in embedding dimension—it isn't just for visualisation! You can use UMAP as a general purpose dimension reduction technique as a preliminary step to other machine learning tasks. With a little care it partners well with the hdbscan clustering library (for more details please see Using UMAP for Clustering).
Third, UMAP often performs better at preserving some aspects of global structure of the data than most implementations of t-SNE. This means that it can often provide a better "big picture" view of your data as well as preserving local neighbor relations.
Fourth, UMAP supports a wide variety of distance functions, including non-metric distance functions such as cosine distance and correlation distance. You can finally embed word vectors properly using cosine distance!
Fifth, UMAP supports adding new points to an existing embedding via the standard sklearn transform method. This means that UMAP can be used as a preprocessing transformer in sklearn pipelines.
Sixth, UMAP supports supervised and semi-supervised dimension reduction. This means that if you have label information that you wish to use as extra information for dimension reduction (even if it is just partial labelling) you can do that—as simply as providing it as the y parameter in the fit method.
Seventh, UMAP supports a variety of additional experimental features including: an "inverse transform" that can approximate a high dimensional sample that would map to a given position in the embedding space; the ability to embed into non-euclidean spaces including hyperbolic embeddings, and embeddings with uncertainty; very preliminary support for embedding dataframes also exists.
Finally, UMAP has solid theoretical foundations in manifold learning (see our paper on ArXiv). This both justifies the approach and allows for further extensions that will soon be added to the library.
Performance and Examples
UMAP is very efficient at embedding large high dimensional datasets. In particular it scales well with both input dimension and embedding dimension. For the best possible performance we recommend installing the nearest neighbor computation library pynndescent . UMAP will work without it, but if installed it will run faster, particularly on multicore machines.
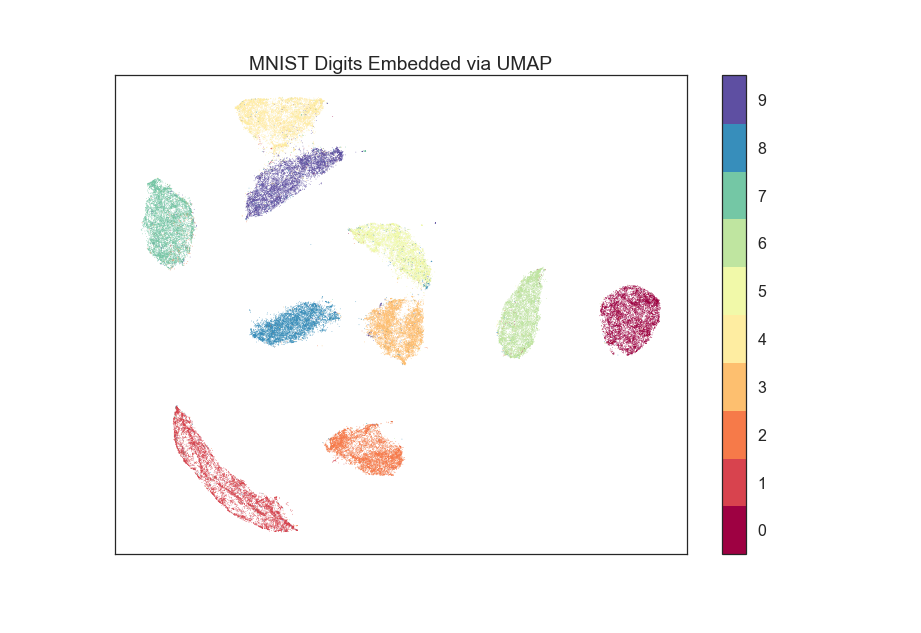
For a problem such as the 784-dimensional MNIST digits dataset with 70000 data samples, UMAP can complete the embedding in under a minute (as compared with around 45 minutes for scikit-learn's t-SNE implementation). Despite this runtime efficiency, UMAP still produces high quality embeddings.
The obligatory MNIST digits dataset, embedded in 42 seconds (with pynndescent installed and after numba jit warmup) using a 3.1 GHz Intel Core i7 processor (n_neighbors=10, min_dist=0.001):
The MNIST digits dataset is fairly straightforward, however. A better test is the more recent "Fashion MNIST" dataset of images of fashion items (again 70000 data sample in 784 dimensions). UMAP produced this embedding in 49 seconds (n_neighbors=5, min_dist=0.1):
The UCI shuttle dataset (43500 sample in 8 dimensions) embeds well under correlation distance in 44 seconds (note the longer time required for correlation distance computations):
The following is a densMAP visualization of the MNIST digits dataset with 784 features based on the same parameters as above (n_neighbors=10, min_dist=0.001). densMAP reveals that the cluster corresponding to digit 1 is noticeably denser, suggesting that there are fewer degrees of freedom in the images of 1 compared to other digits.
Plotting
UMAP includes a subpackage umap.plot for plotting the results of UMAP embeddings. This package needs to be imported separately since it has extra requirements (matplotlib, datashader and holoviews). It allows for fast and simple plotting and attempts to make sensible decisions to avoid overplotting and other pitfalls. An example of use:
import umap
import umap.plot
from sklearn.datasets import load_digits
digits = load_digits()
mapper = umap.UMAP().fit(digits.data)
umap.plot.points(mapper, labels=digits.target)
The plotting package offers basic plots, as well as interactive plots with hover tools and various diagnostic plotting options. See the documentation for more details.
Parametric UMAP
Parametric UMAP provides support for training a neural network to learn a UMAP based transformation of data. This can be used to support faster inference of new unseen data, more robust inverse transforms, autoencoder versions of UMAP and semi-supervised classification (particularly for data well separated by UMAP and very limited amounts of labelled data). See the documentation of Parametric UMAP or the example notebooks for more.
densMAP
The densMAP algorithm augments UMAP to additionally preserve local density information in addition to the topological structure captured by UMAP. One can easily run densMAP using the umap package by setting the densmap input flag:
embedding = umap.UMAP(densmap=True).fit_transform(data)
This functionality is built upon the densMAP implementation provided by the developers of densMAP, who also contributed to integrating densMAP into the umap package.
densMAP inherits all of the parameters of UMAP. The following is a list of additional parameters that can be set for densMAP:
dens_frac: This determines the fraction of epochs (a value between 0 and 1) that will include the density-preservation term in the optimization objective. This parameter is set to 0.3 by default. Note that densMAP switches density optimization on after an initial phase of optimizing the embedding using UMAP.dens_lambda: This determines the weight of the density-preservation objective. Higher values prioritize density preservation, and lower values (closer to zero) prioritize the UMAP objective. Setting this parameter to zero reduces the algorithm to UMAP. Default value is 2.0.dens_var_shift: Regularization term added to the variance of local densities in the embedding for numerical stability. We recommend setting this parameter to 0.1, which consistently works well in many settings.output_dens: When this flag is True, the call tofit_transformreturns, in addition to the embedding, the local radii (inverse measure of local density defined in the densMAP paper) for the original dataset and for the embedding. The output is a tuple(embedding, radii_original, radii_embedding). Note that the radii are log-transformed. If False, only the embedding is returned. This flag can also be used with UMAP to explore the local densities of UMAP embeddings. By default this flag is False.
For densMAP we recommend larger values of n_neighbors (e.g. 30) for reliable estimation of local density.
An example of making use of these options (based on a subsample of the mnist_784 dataset):
import umap
from sklearn.datasets import fetch_openml
from sklearn.utils import resample
digits = fetch_openml(name='mnist_784')
subsample, subsample_labels = resample(digits.data, digits.target, n_samples=7000,
stratify=digits.target, random_state=1)
embedding, r_orig, r_emb = umap.UMAP(densmap=True, dens_lambda=2.0, n_neighbors=30,
output_dens=True).fit_transform(subsample)
See the documentation for more details.
Help and Support
Documentation is at Read the Docs. The documentation includes a FAQ that may answer your questions. If you still have questions then please open an issue and I will try to provide any help and guidance that I can.
Citation
If you make use of this software for your work we would appreciate it if you would cite the paper from the Journal of Open Source Software:
@article{mcinnes2018umap-software,
title={UMAP: Uniform Manifold Approximation and Projection},
author={McInnes, Leland and Healy, John and Saul, Nathaniel and Grossberger, Lukas},
journal={The Journal of Open Source Software},
volume={3},
number={29},
pages={861},
year={2018}
}
If you would like to cite this algorithm in your work the ArXiv paper is the current reference:
@article{2018arXivUMAP,
author = {{McInnes}, L. and {Healy}, J. and {Melville}, J.},
title = "{UMAP: Uniform Manifold Approximation
and Projection for Dimension Reduction}",
journal = {ArXiv e-prints},
archivePrefix = "arXiv",
eprint = {1802.03426},
primaryClass = "stat.ML",
keywords = {Statistics - Machine Learning,
Computer Science - Computational Geometry,
Computer Science - Learning},
year = 2018,
month = feb,
}
Additionally, if you use the densMAP algorithm in your work please cite the following reference:
@article {NBC2020,
author = {Narayan, Ashwin and Berger, Bonnie and Cho, Hyunghoon},
title = {Density-Preserving Data Visualization Unveils Dynamic Patterns of Single-Cell Transcriptomic Variability},
journal = {bioRxiv},
year = {2020},
doi = {10.1101/2020.05.12.077776},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2020/05/14/2020.05.12.077776},
eprint = {https://www.biorxiv.org/content/early/2020/05/14/2020.05.12.077776.full.pdf},
}
If you use the Parametric UMAP algorithm in your work please cite the following reference:
@article {NBC2020,
author = {Sainburg, Tim and McInnes, Leland and Gentner, Timothy Q.},
title = {Parametric UMAP: learning embeddings with deep neural networks for representation and semi-supervised learning},
journal = {ArXiv e-prints},
archivePrefix = "arXiv",
eprint = {},
primaryClass = "stat.ML",
keywords = {Statistics - Machine Learning,
Computer Science - Computational Geometry,
Computer Science - Learning},
year = 2020,
}
License
The umap package is 3-clause BSD licensed.
We would like to note that the umap package makes heavy use of NumFOCUS sponsored projects, and would not be possible without their support of those projects, so please consider contributing to NumFOCUS.
Contributing
Contributions are more than welcome! There are lots of opportunities for potential projects, so please get in touch if you would like to help out. Everything from code to notebooks to examples and documentation are all equally valuable so please don't feel you can't contribute. To contribute please fork the project make your changes and submit a pull request. We will do our best to work through any issues with you and get your code merged into the main branch.