Welcome to
Official repository of deepflash2 - a deep-learning pipeline for segmentation of ambiguous microscopic images.
Quick Start in 30 seconds
setup5.mov
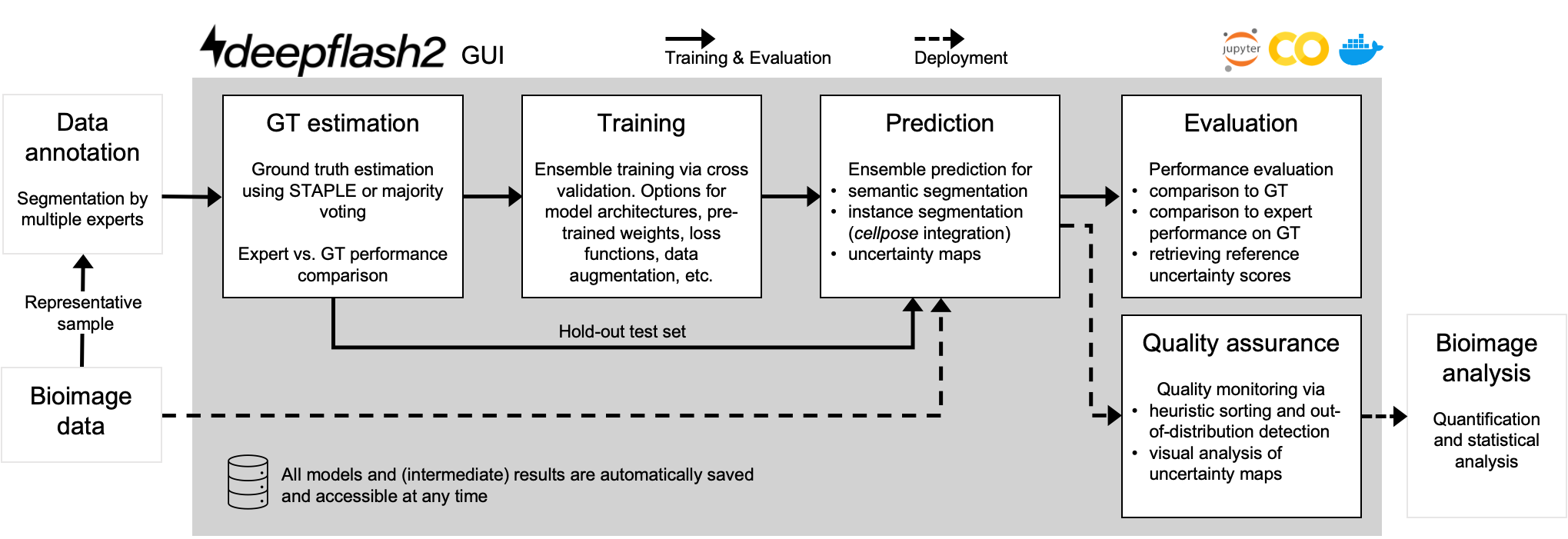
Why using deepflash2?
The best of two worlds: Combining state of the art deep learning with a barrier free environment for life science researchers.
- End-to-end process for life science researchers
- graphical user interface - no coding skills required
- free usage on Google Colab at no costs
- easy deployment on own hardware
- Reliable prediction on new data
- Quality assurance and out-of-distribution detection
Kaggle Gold Medal and Innovation Price Winner
deepflash2 does not only work on fluorescent labels. The deepflash2 API built the foundation for winning the Innovation Award a Kaggle Gold Medal in the HuBMAP - Hacking the Kidney challenge. Have a look at our solution
Citing
The preprint of our paper is available on arXiv. Please cite
@misc{griebel2021deepflash2,
title={Deep-learning in the bioimaging wild: Handling ambiguous data with deepflash2},
author={Matthias Griebel and Dennis Segebarth and Nikolai Stein and Nina Schukraft and Philip Tovote and Robert Blum and Christoph M. Flath},
year={2021},
eprint={2111.06693}
}
Installing
You can use deepflash2 by using Google Colab. You can run every page of the documentation as an interactive notebook - click "Open in Colab" at the top of any page to open it.
- Be sure to change the Colab runtime to "GPU" to have it run fast!
- Use Firefox or Google Chrome if you want to upload your images.
You can install deepflash2 on your own machines with conda (highly recommended):
conda install -c fastchan -c matjesg deepflash2
To install with pip, use
pip install deepflash2
If you install with pip, you should install PyTorch first by following the installation instructions of pytorch or fastai.
Using Docker
Docker images for deepflash2 are built on top of the latest pytorch image and fastai images. You must install Nvidia-Docker to enable gpu compatibility with these containers.
- CPU only
docker run -p 8888:8888 matjesg/deepflash
- With GPU support (Nvidia-Docker must be installed.) has an editable install of fastai and fastcore.
docker run --gpus all -p 8888:8888 matjesg/deepflashAll docker containers are configured to start a jupyter server. deepflash2 notebooks are available in thedeepflash2_notebooksfolder.
For more information on how to run docker see docker orientation and setup and fastai docker.
Creating segmentation masks with Fiji/ImageJ
If you don't have labelled training data available, you can use this instruction manual for creating segmentation maps. The ImagJ-Macro is available here.
Acronym
A Deep-learning pipeline for Fluorescent Label Segmentation that learns from Human experts