12 Repositories
Python sequencing Libraries
Analysis of Antarctica sequencing samples contaminated with SARS-CoV-2
Analysis of SARS-CoV-2 reads in sequencing of 2018-2019 Antarctica samples in PRJNA692319 The samples analyzed here are described in this preprint, wh

Sequence lineage information extracted from RKI sequence data repo
Pango lineage information for German SARS-CoV-2 sequences This repository contains a join of the metadata and pango lineage tables of all German SARS-
dragmap-meth: Fast and accurate aligner for bisulfite sequencing reads using dragmap
dragmap_meth (dragmap_meth.py) Alignment of BS-Seq reads using dragmap. Intro This works for single-end reads and for paired-end reads from the direct
ONT Analysis Toolkit (OAT)
A toolkit for monitoring ONT MinION sequencing, followed by data analysis, for viral genomes amplified with tiled amplicon sequencing.
ScisorWiz: Differential Isoform Visualizer for Long-Read RNA Sequencing Data
ScisorWiz: Vizualizer for Differential Isoform Expression README ScisorWiz is a linux-based R-package for visualizing differential isoform expression
Cash in on Expressed Barcode Tags (EBTs) from NGS Sequencing Data with Python
Cash in on Expressed Barcode Tags (EBTs) from NGS Sequencing Data with Python Cashier is a tool developed by Russell Durrett for the analysis and extr
HTSeq is a Python library to facilitate processing and analysis of data from high-throughput sequencing (HTS) experiments.
HTSeq DEVS: https://github.com/htseq/htseq DOCS: https://htseq.readthedocs.io A Python library to facilitate programmatic analysis of data from high-t
MHtyper is an end-to-end pipeline for recognized the Forensic microhaplotypes in Nanopore sequencing data.
MHtyper is an end-to-end pipeline for recognized the Forensic microhaplotypes in Nanopore sequencing data. It is implemented using Python.

cLoops2: full stack analysis tool for chromatin interactions
cLoops2: full stack analysis tool for chromatin interactions Introduction cLoops2 is an extension of our previous work, cLoops. From loop-calling base
InDels analysis of CRISPR lines by NGS amplicon sequencing technology for a multicopy gene family.
CRISPRanalysis InDels analysis of CRISPR lines by NGS amplicon sequencing technology for a multicopy gene family. In this work, we present a workflow
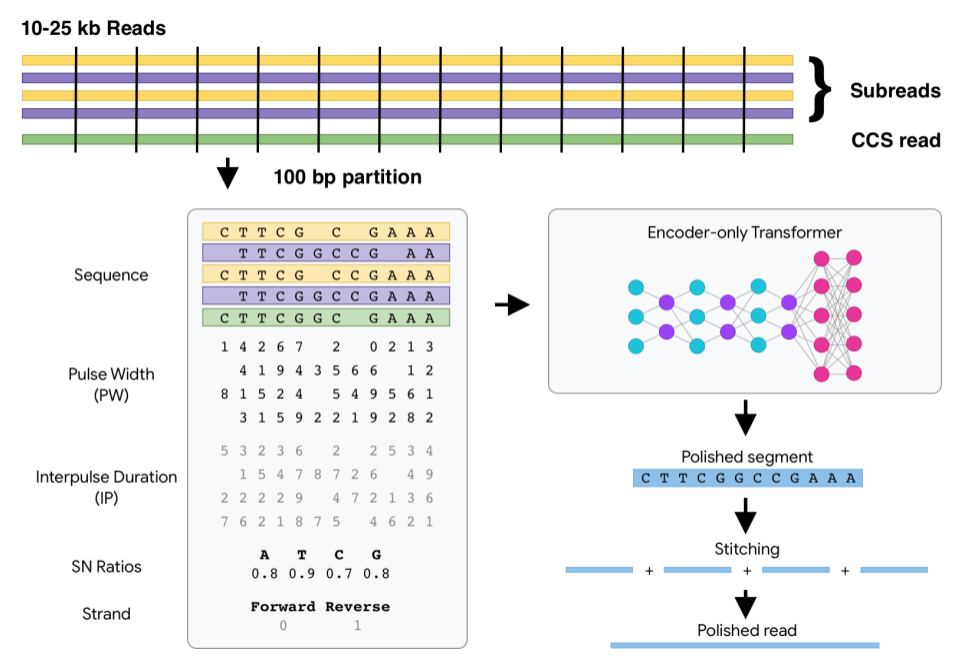
DeepConsensus uses gap-aware sequence transformers to correct errors in Pacific Biosciences (PacBio) Circular Consensus Sequencing (CCS) data.
DeepConsensus DeepConsensus uses gap-aware sequence transformers to correct errors in Pacific Biosciences (PacBio) Circular Consensus Sequencing (CCS)
Data intensive science for everyone.
The latest information about Galaxy can be found on the Galaxy Community Hub. Community support is available at Galaxy Help. Galaxy Quickstart Galaxy