PROTEIN-EXPRESSION-ANALYSIS-FOR-DOWN-SYNDROME
Down syndrome (DS) is a chromosomal disorder where organisms have an extra chromosome 21, sometimes known as trisomy 21. Being a syndrome, DS consists of multiple symptoms affecting a large number of systems in the body. It’s effect on learning results in it being an intellectual disability . Memantine is currently proposed as a treatment of the learning deficit symptoms in DS. In this project, we have used several supervised machine learning methods:logistic regression, a random forest classifier,k nearest neighbour and voting classifier, to identify which protein(s) are critical to mice learning ability after being exposed to context fear conditioning (CFC). 77 protein expression levels are analysed from both control and trisomic (Ts65Dn) genotype mice, both with and without treatment from the drug memantine. Results suggest that voting classification approach can identify the most important proteins which may help to identify more effective drugs to help the learning process in people with DS.
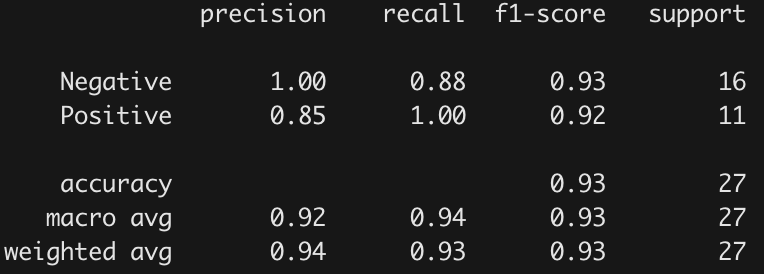
Identified proteins critical to learning ability and classification of mice based on protein expression values using machine learning algorithms.