51 Repositories
Python bioinformatics Libraries

Precision Medicine Knowledge Graph (PrimeKG)
PrimeKG Website | bioRxiv Paper | Harvard Dataverse Precision Medicine Knowledge Graph (PrimeKG) presents a holistic view of diseases. PrimeKG integra

MarcoPolo is a clustering-free approach to the exploration of bimodally expressed genes along with group information in single-cell RNA-seq data
MarcoPolo is a method to discover differentially expressed genes in single-cell RNA-seq data without depending on prior clustering Overview MarcoPolo
A sage package for working with circular genomes represented by signed or unsigned permutations
Circular genome tools (cgt) A sage package for working with circular genomes represented by signed or unsigned permutations. It includes tools for con
NFCDS Workshop Beginners Guide Bioinformatics Data Analysis
Genomics Workshop FIXME: overview of workshop Code of Conduct All participants s

Reverse engineering the dengue virus (under development construction)
Reverse engineering the dengue virus (under development 🚧 ) What is dengue? Dengue is a viral infection transmitted to humans through the bite of inf

NNR conformation conditional and global probabilities estimation and analysis in peptides or proteins fragments
NNR and global probabilities estimation and analysis in peptides or protein fragments This module calculates global and NNR conformation dependent pro
Pca-on-genotypes - Mini bioinformatics project - PCA on genotypes
Mini bioinformatics project: PCA on genotypes This repo contains the code from t
NeoDTI: Neural integration of neighbor information from a heterogeneous network for discovering new drug-target interactions
NeoDTI NeoDTI: Neural integration of neighbor information from a heterogeneous network for discovering new drug-target interactions (Bioinformatics).
Exam assignment for Laboratory of Bioinformatics 2
Exam assignment for Laboratory of Bioinformatics 2 (Alma Mater University of Bologna, Master in Bioinformatics)

Retrieve annotated intron sequences and classify them as minor (U12-type) or major (U2-type)
(intron I nterrogator and C lassifier) intronIC is a program that can be used to classify intron sequences as minor (U12-type) or major (U2-type), usi

BisQue is a web-based platform designed to provide researchers with organizational and quantitative analysis tools for 5D image data. Users can extend BisQue by implementing containerized ML workflows.
Overview BisQue is a web-based platform specifically designed to provide researchers with organizational and quantitative analysis tools for up to 5D

A web-based app that allows easy, simple - and if desired high-throughput - analysis of qPCR data
qpcr-Analyser A web-based GUI for the qpcr package that allows easy, simple and high-throughput analysis of qPCR data. As is described in more detail
Simple web browser to visualize HiC tracks
HiCBrowser : A simple web browser to visualize Hi-C and other genomic tracks Fidel Ramirez, José Villaveces, Vivek Bhardwaj Installation You can insta

Full Spectrum Bioinformatics - a free online text designed to introduce key topics in Bioinformatics using the Python
Full Spectrum Bioinformatics is a free online text designed to introduce key topics in Bioinformatics using the Python programming language. The text is written in interactive Jupyter Notebooks, which allow you to try out and modify example code and analyses.

PyIOmica (pyiomica) is a Python package for omics analyses.
PyIOmica (pyiomica) This repository contains PyIOmica, a Python package that provides bioinformatics utilities for analyzing (dynamic) omics datasets.
Programming in Bioinformatics, Block 3
Programming in Bioinformatics - Block 3 I. Setting up Environment and Running the Code Create the environment using the pibi_block3.yml file with the
Sequence clustering and database creation using mmseqs, from local fasta files
Sequence clustering and database creation using mmseqs, from local fasta files

A DSL for data-driven computational pipelines
"Dataflow variables are spectacularly expressive in concurrent programming" Henri E. Bal , Jennifer G. Steiner , Andrew S. Tanenbaum Quick overview Ne
Single-Cell Analysis in Python. Scales to 1M cells.
Scanpy – Single-Cell Analysis in Python Scanpy is a scalable toolkit for analyzing single-cell gene expression data built jointly with anndata. It inc
Collapse a set of redundant kmers to use IUPAC degenerate bases
kmer-collapse Collapse a set of redundant kmers to use IUPAC degenerate bases Overview Given an input set of kmers, find the smallest set of kmers tha
AptaMat is a simple script which aims to measure differences between DNA or RNA secondary structures.
AptaMAT Purpose AptaMat is a simple script which aims to measure differences between DNA or RNA secondary structures. The method is based on the compa
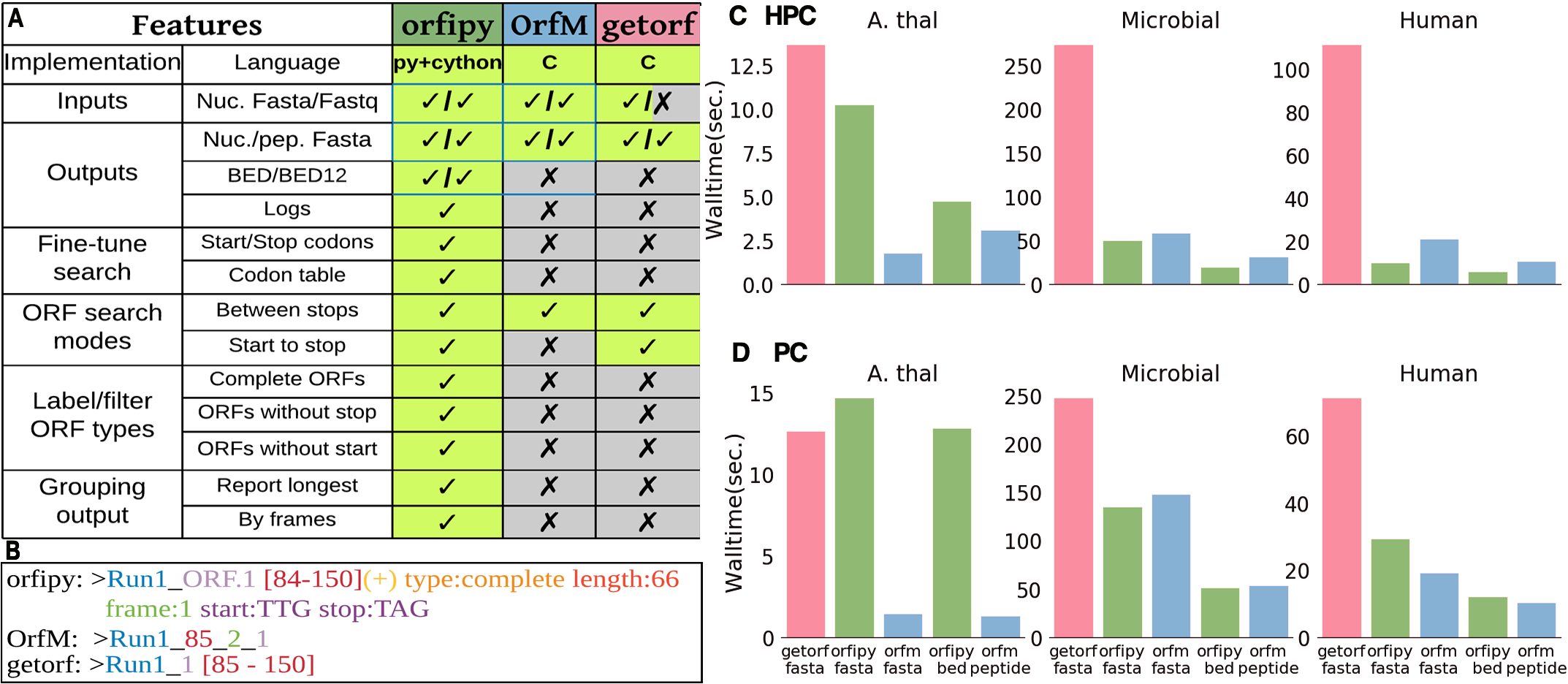
orfipy is a tool written in python/cython to extract ORFs in an extremely and fast and flexible manner
Introduction orfipy is a tool written in python/cython to extract ORFs in an extremely and fast and flexible manner. Other popular ORF searching tools
A tool to visualise the results of AlphaFold2 and inspect the quality of structural predictions
AlphaFold Analyser This program produces high quality visualisations of predicted structures produced by AlphaFold. These visualisations allow the use

cLoops2: full stack analysis tool for chromatin interactions
cLoops2: full stack analysis tool for chromatin interactions Introduction cLoops2 is an extension of our previous work, cLoops. From loop-calling base

Bioinformatics tool for exploring RNA-Protein interactions
Explore RNA-Protein interactions. RNPFind is a bioinformatics tool. It takes an RNA transcript as input and gives a list of RNA binding protein (RBP)
Michael Vinyard's utilities
Install vintools To download this package from pypi: pip install vintools Install the development package To download and install the developmen
ToeholdTools is a Python package and desktop app designed to facilitate analyzing and designing toehold switches, created as part of the 2021 iGEM competition.
ToeholdTools Category Status Repository Package Build Quality A library for the analysis of toehold switch riboregulators created by the iGEM team Cit
Algorithms covered in the Bioinformatics Course part of the Cambridge Computer Science Tripos
Bioinformatics This is a repository of all the algorithms covered in the Bioinformatics Course part of the Cambridge Computer Science Tripos Algorithm
Common bioinformatics database construction
biodb Common bioinformatics database construction 1.taxonomy (Substance classification database) Download the database wget -c https://ftp.ncbi.nlm.ni
Tool for working with Y-chromosome data from YFull and FTDNA
ycomp ycomp is a tool for working with Y-chromosome data from YFull and FTDNA. Run ycomp -h for information on how to use the program. Installation Th
A Snakemake workflow for standardised sc/snRNAseq analysis
single_snake_sequencing - sc/snRNAseq Snakemake Workflow A Snakemake workflow for standardised sc/snRNAseq analysis. Every single cell analysis is sli
Toolchest provides APIs for scientific and bioinformatic data analysis.
Toolchest Python Client Toolchest provides APIs for scientific and bioinformatic data analysis. It allows you to abstract away the costliness of runni
A geometric deep learning pipeline for predicting protein interface contacts.
A geometric deep learning pipeline for predicting protein interface contacts.

pcnaDeep integrates cutting-edge detection techniques with tracking and cell cycle resolving models.
pcnaDeep: a deep-learning based single-cell cycle profiler with PCNA signal Welcome! pcnaDeep integrates cutting-edge detection techniques with tracki

Implementation of Neural Distance Embeddings for Biological Sequences (NeuroSEED) in PyTorch
Neural Distance Embeddings for Biological Sequences Official implementation of Neural Distance Embeddings for Biological Sequences (NeuroSEED) in PyTo
Cool Bioinformatics Scripts
Cool Bioinformatics Scripts qqplot You can use this script in two ways read tons of millions of P values from stdin # python zcat pval.txt.gz | qqplo
An adaptable Snakemake workflow which uses GATKs best practice recommendations to perform germline mutation calling starting with BAM files
Germline Mutation Calling This Snakemake workflow follows the GATK best-practice recommandations to call small germline variants. The pipeline require
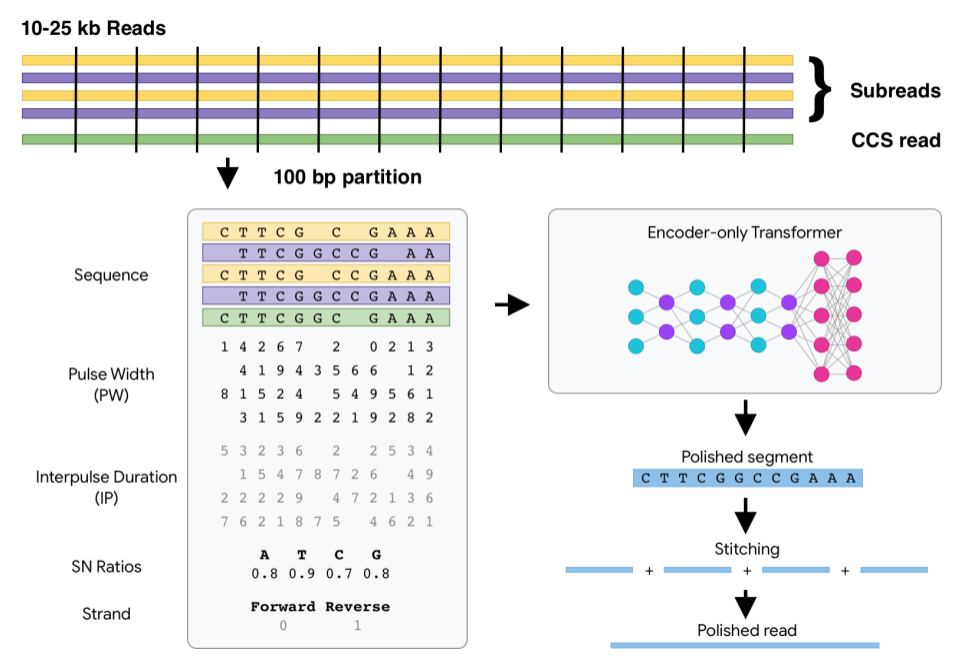
DeepConsensus uses gap-aware sequence transformers to correct errors in Pacific Biosciences (PacBio) Circular Consensus Sequencing (CCS) data.
DeepConsensus DeepConsensus uses gap-aware sequence transformers to correct errors in Pacific Biosciences (PacBio) Circular Consensus Sequencing (CCS)
Main repository for the HackBio'2021 Virtual Internship Experience for #Team-Greider ❤️
Hello 🤟 #Team-Greider The team of 20 people for HackBio'2021 Virtual Bioinformatics Internship 💝 🖨️ 👨💻 HackBio: https://thehackbio.com 💬 Ask us

Drug design and development team HackBio internship is a virtual bioinformatics program that introduces students and professional to advanced practical bioinformatics and its applications globally.
-Nyokong. Drug design and development team HackBio internship is a virtual bioinformatics program that introduces students and professional to advance

TeamFleming is a multicultural group of 20 young bioinformatics enthusiasts participating in the 2021 HackBio Virtual Summer Internship
💻 Welcome to Team Fleming's Repo! #TeamFleming is a multicultural group of 20 young bioinformatics enthusiasts participating in the 2021 HackBio Virt
This is the repo for the paper `SumGNN: Multi-typed Drug Interaction Prediction via Efficient Knowledge Graph Summarization'. (published in Bioinformatics'21)
SumGNN: Multi-typed Drug Interaction Prediction via Efficient Knowledge Graph Summarization This is the code for our paper ``SumGNN: Multi-typed Drug
Incubator for useful bioinformatics code, primarily in Python and R
Collection of useful code related to biological analysis. Much of this is discussed with examples at Blue collar bioinformatics. All code, images and

Request execution of Galaxy SARS-CoV-2 variation analysis workflows on input data you provide.
SARS-CoV-2 processing requests Request execution of Galaxy SARS-CoV-2 variation analysis workflows on input data you provide. Prerequisites This autom
Data intensive science for everyone.
The latest information about Galaxy can be found on the Galaxy Community Hub. Community support is available at Galaxy Help. Galaxy Quickstart Galaxy

A full spaCy pipeline and models for scientific/biomedical documents.
This repository contains custom pipes and models related to using spaCy for scientific documents. In particular, there is a custom tokenizer that adds

Analytical Web Apps for Python, R, Julia, and Jupyter. No JavaScript Required.
Dash Dash is the most downloaded, trusted Python framework for building ML & data science web apps. Built on top of Plotly.js, React and Flask, Dash t

A full spaCy pipeline and models for scientific/biomedical documents.
This repository contains custom pipes and models related to using spaCy for scientific documents. In particular, there is a custom tokenizer that adds

Analytical Web Apps for Python, R, Julia, and Jupyter. No JavaScript Required.
Dash Dash is the most downloaded, trusted Python framework for building ML & data science web apps. Built on top of Plotly.js, React and Flask, Dash t
Incubator for useful bioinformatics code, primarily in Python and R
Collection of useful code related to biological analysis. Much of this is discussed with examples at Blue collar bioinformatics. All code, images and

Analytical Web Apps for Python, R, Julia, and Jupyter. No JavaScript Required.
Dash Dash is the most downloaded, trusted Python framework for building ML & data science web apps. Built on top of Plotly.js, React and Flask, Dash t