19 Repositories
Python molecules Libraries
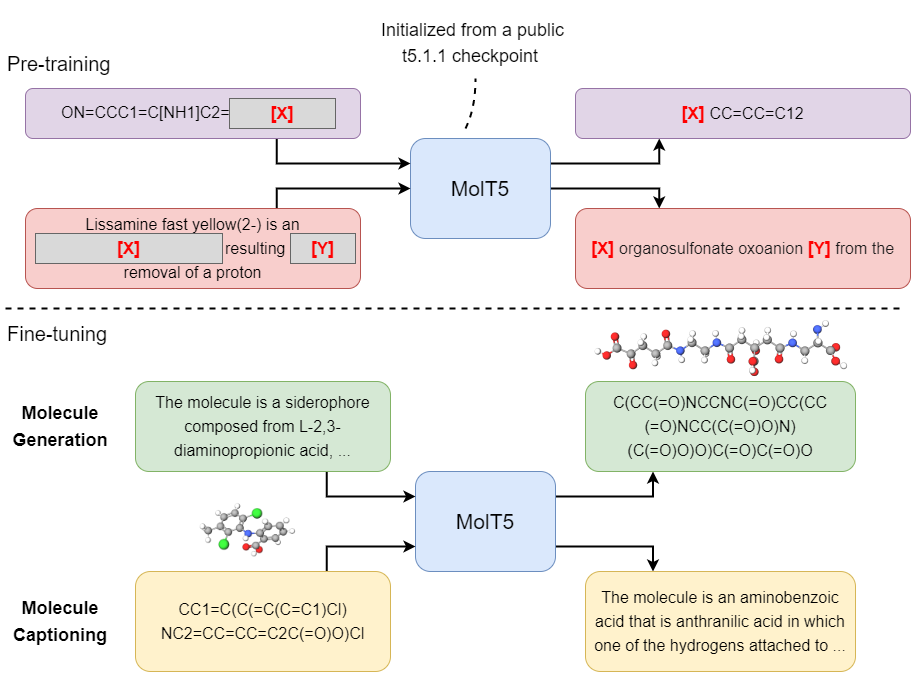
Associated Repository for "Translation between Molecules and Natural Language"
MolT5: Translation between Molecules and Natural Language Associated repository for "Translation between Molecules and Natural Language". Table of Con

Official implementation of "Generating 3D Molecules for Target Protein Binding"
Generating 3D Molecules for Target Protein Binding This is the official implementation of the GraphBP method proposed in the following paper. Meng Liu

EquiBind: Geometric Deep Learning for Drug Binding Structure Prediction
EquiBind: geometric deep learning for fast predictions of the 3D structure in which a small molecule binds to a protein

A basic molecule viewer written in Python, using curses; Thus, meant for linux terminals
asciiMOL A basic molecule viewer written in Python, using curses; Thus, meant for linux terminals. This is an alpha version, featuring: Opening defaul
E2EDNA2 - An automated pipeline for simulation of DNA aptamers complexed with small molecules and short peptides
E2EDNA2 - An automated pipeline for simulation of DNA aptamers complexed with small molecules and short peptides

A short and easy PyTorch implementation of E(n) Equivariant Graph Neural Networks
Simple implementation of Equivariant GNN A short implementation of E(n) Equivariant Graph Neural Networks for HOMO energy prediction. Just 50 lines of

Splore - a simple graphical interface for scrolling through and exploring data sets of molecules
Scroll through and exPLORE molecule sets The splore framework aims to offer a si
A fast Protein Chain / Ligand Extractor and organizer.
Are you tired of using visualization software, or full blown suites just to separate protein chains / ligands ? Are you tired of organizing the mess o

Interactive chemical viewer for 2D structures of small molecules
👀 mols2grid mols2grid is an interactive chemical viewer for 2D structures of small molecules, based on RDKit. ➡️ Try the demo notebook on Google Cola
GemNet model in PyTorch, as proposed in "GemNet: Universal Directional Graph Neural Networks for Molecules" (NeurIPS 2021)
GemNet: Universal Directional Graph Neural Networks for Molecules Reference implementation in PyTorch of the geometric message passing neural network
FS-Mol: A Few-Shot Learning Dataset of Molecules
FS-Mol is A Few-Shot Learning Dataset of Molecules, containing molecular compounds with measurements of activity against a variety of protein targets. The dataset is presented with a model evaluation benchmark which aims to drive few-shot learning research in the domain of molecules and graph-structured data.
Python module for drawing and rendering beautiful atoms and molecules using Blender.
Batoms is a Python package for editing and rendering atoms and molecules objects using blender. A Python interface that allows for automating workflows.

Source code for GNN-LSPE (Graph Neural Networks with Learnable Structural and Positional Representations)
Graph Neural Networks with Learnable Structural and Positional Representations Source code for the paper "Graph Neural Networks with Learnable Structu
Codebase of deep learning models for inferring stability of mRNA molecules
Kaggle OpenVaccine Models Codebase of deep learning models for inferring stability of mRNA molecules, corresponding to the Kaggle Open Vaccine Challen

Making self-supervised learning work on molecules by using their 3D geometry to pre-train GNNs. Implemented in DGL and Pytorch Geometric.
3D Infomax improves GNNs for Molecular Property Prediction Video | Paper We pre-train GNNs to understand the geometry of molecules given only their 2D
Library for processing molecules and reactions in python way
Chython [ˈkʌɪθ(ə)n] Library for processing molecules and reactions in python way. Features: Read/write/convert formats: MDL .RDF (.RXN) and .SDF (.MOL

Datamol is a python library to work with molecules
Datamol is a python library to work with molecules. It's a layer built on top of RDKit and aims to be as light as possible.

Datamol is a python library to work with molecules.
Datamol is a python library to work with molecules. It's a layer built on top of RDKit and aims to be as light as possible.

A PyTorch implementation of "Pathfinder Discovery Networks for Neural Message Passing"
A PyTorch implementation of "Pathfinder Discovery Networks for Neural Message Passing" (WebConf 2021). Abstract In this work we propose Pathfind