22 Repositories
Python circular-genome Libraries
A spatial genome aligner for analyzing multiplexed DNA-FISH imaging data.
jie jie is a spatial genome aligner. This package parses true chromatin imaging signal from noise by aligning signals to a reference DNA polymer model
A sage package for working with circular genomes represented by signed or unsigned permutations
Circular genome tools (cgt) A sage package for working with circular genomes represented by signed or unsigned permutations. It includes tools for con
Gapmm2: gapped alignment using minimap2 (align transcripts to genome)
gapmm2: gapped alignment using minimap2 This tool is a wrapper for minimap2 to r
Pipeline code for Sequential-GAM(Genome Architecture Mapping).
Sequential-GAM Pipeline code for Sequential-GAM(Genome Architecture Mapping). mapping whole_preprocess.sh include the whole processing of mapping. usa
A High-Level Fusion Scheme for Circular Quantities published at the 20th International Conference on Advanced Robotics
Monte Carlo Simulation to the Paper A High-Level Fusion Scheme for Circular Quantities published at the 20th International Conference on Advanced Robotics

Classification of Long Sequential Data using Circular Dilated Convolutional Neural Networks
Classification of Long Sequential Data using Circular Dilated Convolutional Neural Networks arXiv preprint: https://arxiv.org/abs/2201.02143. Architec

Train Scene Graph Generation for Visual Genome and GQA in PyTorch = 1.2 with improved zero and few-shot generalization.
Scene Graph Generation Object Detections Ground truth Scene Graph Generated Scene Graph In this visualization, woman sitting on rock is a zero-shot tr

Bottom-up attention model for image captioning and VQA, based on Faster R-CNN and Visual Genome
bottom-up-attention This code implements a bottom-up attention model, based on multi-gpu training of Faster R-CNN with ResNet-101, using object and at

Retrieve annotated intron sequences and classify them as minor (U12-type) or major (U2-type)
(intron I nterrogator and C lassifier) intronIC is a program that can be used to classify intron sequences as minor (U12-type) or major (U2-type), usi
These are the scripts used for the project of ‘Assembly of a pan-genome for global cattle reveals missing sequence and novel structural variation, providing new insights into their diversity and evolution history’
script-SV-genotyping These are the scripts used for the project of ‘Assembly of a pan-genome for global cattle reveals missing sequence and novel stru

Python library to visualize circular plasmid maps
Plasmidviewer Plasmidviewer is a Python library to visualize plasmid maps from GenBank. This library provides only the function to visualize circular
Generate "Jupiter" plots for circular genomes
jupiter Generate "Jupiter" plots for circular genomes Description Python scripts to generate plots from ViennaRNA output. Written in "pidgin" python w
Replace sequence_IDs in gff3 based on given genome.fasta
gff-rename Replace the sequence IDs in a gff3 file with a set of provided sequence IDs from a genom.fasta. This is useful when a gff3 file is retrieve

cLoops2: full stack analysis tool for chromatin interactions
cLoops2: full stack analysis tool for chromatin interactions Introduction cLoops2 is an extension of our previous work, cLoops. From loop-calling base
CBMPy Metadraft: a flexible and extensible genome-scale model reconstruction tool
CBMPy Metadraft: a flexible and extensible, GUI-based genome-scale model reconstruction tool that supports multiple Systems Biology standards.
Identify and annotate mutations from genome editing assays.
CRISPR-detector Here we propose our CRISPR-detector to facilitate the CRISPR-edited amplicon and whole genome sequencing data analysis, with functions
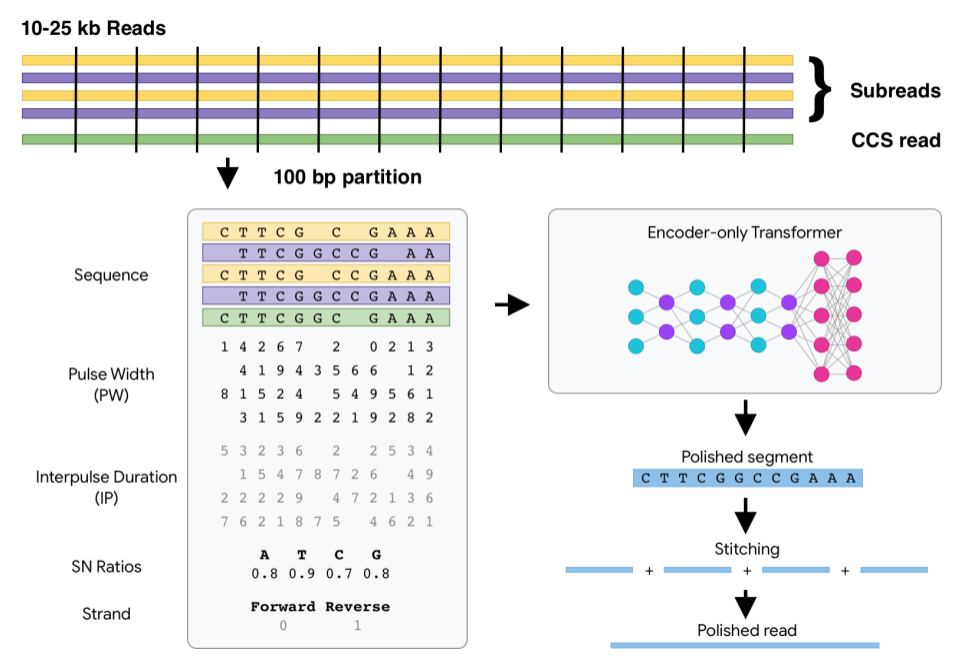
DeepConsensus uses gap-aware sequence transformers to correct errors in Pacific Biosciences (PacBio) Circular Consensus Sequencing (CCS) data.
DeepConsensus DeepConsensus uses gap-aware sequence transformers to correct errors in Pacific Biosciences (PacBio) Circular Consensus Sequencing (CCS)

Code for "Learning Canonical Representations for Scene Graph to Image Generation", Herzig & Bar et al., ECCV2020
Learning Canonical Representations for Scene Graph to Image Generation (ECCV 2020) Roei Herzig*, Amir Bar*, Huijuan Xu, Gal Chechik, Trevor Darrell, A
minimizer-space de Bruijn graphs (mdBG) for whole genome assembly
rust-mdbg: Minimizer-space de Bruijn graphs (mdBG) for whole-genome assembly rust-mdbg is an ultra-fast minimizer-space de Bruijn graph (mdBG) impleme

Statistical package in Python based on Pandas
Pingouin is an open-source statistical package written in Python 3 and based mostly on Pandas and NumPy. Some of its main features are listed below. F
To be a next-generation DL-based phenotype prediction from genome mutations.
Sequence -----------+-- 3D_structure -- 3D_module --+ +-- ? | |
Tool for pinpointing circular imports in Python. Find cyclic imports in any project
Pycycle: Find and fix circular imports in python projects Pycycle is an experimental project that aims to help python developers fix their circular de