50 Repositories
Python molecular Libraries

Jupyter Dock is a set of Jupyter Notebooks for performing molecular docking protocols interactively, as well as visualizing, converting file formats and analyzing the results.
Molecular Docking integrated in Jupyter Notebooks Description | Citation | Installation | Examples | Limitations | License Table of content Descriptio

Implementation of GeoDiff: a Geometric Diffusion Model for Molecular Conformation Generation (ICLR 2022).
GeoDiff: a Geometric Diffusion Model for Molecular Conformation Generation [OpenReview] [arXiv] [Code] The official implementation of GeoDiff: A Geome

Addon and nodes for working with structural biology and molecular data in Blender.
Molecular Nodes 🧬 🔬 💻 Buy Me a Coffee to Keep Development Going! Join a Community of Blender SciVis People! What is Molecular Nodes? Molecular Node

TorchMD-Net provides state-of-the-art graph neural networks and equivariant transformer neural networks potentials for learning molecular potentials
TorchMD-net TorchMD-Net provides state-of-the-art graph neural networks and equivariant transformer neural networks potentials for learning molecular
This repository contains the code for Direct Molecular Conformation Generation (DMCG).
Direct Molecular Conformation Generation This repository contains the code for Direct Molecular Conformation Generation (DMCG). Dataset Download rdkit

Chem: collection of mostly python code for molecular visualization, QM/MM, FEP, etc
chem: collection of mostly python code for molecular visualization, QM/MM, FEP,
MoBioTools A simple yet versatile toolkit to automatically setup quantum mechanics/molecular mechanics
A simple yet versatile toolkit to setup quantum mechanical/molecular mechanical (QM/MM) calculations from molecular dynamics trajectories.

Boltzmann visualization - Visualize the Boltzmann distribution for simple quantum models of molecular motion
Boltzmann visualization - Visualize the Boltzmann distribution for simple quantum models of molecular motion

TUPÃ was developed to analyze electric field properties in molecular simulations
TUPÃ: Electric field analyses for molecular simulations What is TUPÃ? TUPÃ (pronounced as tu-pan) is a python algorithm that employs MDAnalysis engine
Riemannian Geometry for Molecular Surface Approximation (RGMolSA)
Riemannian Geometry for Molecular Surface Approximation (RGMolSA) Introduction Ligand-based virtual screening aims to reduce the cost and duration of
Python implementation of the multistate Bennett acceptance ratio (MBAR)
pymbar Python implementation of the multistate Bennett acceptance ratio (MBAR) method for estimating expectations and free energy differences from equ

Albert launcher extension for converting units of length, mass, speed, temperature, time, current, luminosity, printing measurements, molecular substance, and more
unit-converter-albert-ext Extension for converting units of length, mass, speed, temperature, time, current, luminosity, printing measurements, molecu
A pythonic interface to high-throughput virtual screening software
pyscreener A pythonic interface to high-throughput virtual screening software Overview This repository contains the source of pyscreener, both a libra
Official code for the publication "HyFactor: Hydrogen-count labelled graph-based defactorization Autoencoder".
HyFactor Graph-based architectures are becoming increasingly popular as a tool for structure generation. Here, we introduce a novel open-source archit
MDAnalysis is a Python library to analyze molecular dynamics simulations.
MDAnalysis Repository README [*] MDAnalysis is a Python library for the analysis of computer simulations of many-body systems at the molecular scale,
GeoMol: Torsional Geometric Generation of Molecular 3D Conformer Ensembles
GeoMol: Torsional Geometric Generation of Molecular 3D Conformer Ensembles This repository contains a method to generate 3D conformer ensembles direct
Rapid experimentation and scaling of deep learning models on molecular and crystal graphs.
LitMatter A template for rapid experimentation and scaling deep learning models on molecular and crystal graphs. How to use Clone this repository and
Junction Tree Variational Autoencoder for Molecular Graph Generation (ICML 2018)
Junction Tree Variational Autoencoder for Molecular Graph Generation Official implementation of our Junction Tree Variational Autoencoder https://arxi
UF3: a python library for generating ultra-fast interatomic potentials
Ultra-Fast Force Fields (UF3) S. R. Xie, M. Rupp, and R. G. Hennig, "Ultra-fast interpretable machine-learning potentials", preprint arXiv:2110.00624

Source code for our paper "Molecular Mechanics-Driven Graph Neural Network with Multiplex Graph for Molecular Structures"
Molecular Mechanics-Driven Graph Neural Network with Multiplex Graph for Molecular Structures Code for the Multiplex Molecular Graph Neural Network (M
![Code and Data for the paper: Molecular Contrastive Learning with Chemical Element Knowledge Graph [AAAI 2022]](https://github.com/Fangyin1994/KCL/raw/main/fig/overview.png)
Code and Data for the paper: Molecular Contrastive Learning with Chemical Element Knowledge Graph [AAAI 2022]
Knowledge-enhanced Contrastive Learning (KCL) Molecular Contrastive Learning with Chemical Element Knowledge Graph [ AAAI 2022 ]. We construct a Chemi
An automatic reaction network generator for reactive molecular dynamics simulation.
ReacNetGenerator An automatic reaction network generator for reactive molecular dynamics simulation. ReacNetGenerator: an automatic reaction network g

🥇Samsung AI Challenge 2021 1등 솔루션입니다🥇
MoT - Molecular Transformer Large-scale Pretraining for Molecular Property Prediction Samsung AI Challenge for Scientific Discovery This repository is
Small wrapper around 3dmol.js and html2canvas for creating self-contained HTML files that display a 3D molecular representation.
Description Small wrapper around 3dmol.js and html2canvas for creating self-contained HTML files that display a 3D molecular representation. Double cl

Molecular Sets (MOSES): A Benchmarking Platform for Molecular Generation Models
Molecular Sets (MOSES): A benchmarking platform for molecular generation models Deep generative models are rapidly becoming popular for the discovery
CINECA molecular dynamics tutorial set
High Performance Molecular Dynamics Logging into CINECA's computer systems To logon to the M100 system use the following command from an SSH client ss
Powerful, efficient particle trajectory analysis in scientific Python.
freud Overview The freud Python library provides a simple, flexible, powerful set of tools for analyzing trajectories obtained from molecular dynamics

This package is a python library with tools for the Molecular Simulation - Software Gromos.
This package is a python library with tools for the Molecular Simulation - Software Gromos. It allows you to easily set up, manage and analyze simulations in python.
A primitive Python wrapper around the Gromacs tools.
README: GromacsWrapper A primitive Python wrapper around the Gromacs tools. The library is tested with GROMACS 4.6.5, 2018.x, 2019.x, 2020.x, and 2021

Animate molecular orbital transitions using Psi4 and Blender
Molecular Orbital Transitions (MOT) Animate molecular orbital transitions using Psi4 and Blender Author: Maximilian Paradiz Dominguez, University of A

Molecular Sets (MOSES): A benchmarking platform for molecular generation models
Molecular Sets (MOSES): A benchmarking platform for molecular generation models Deep generative models are rapidly becoming popular for the discovery

Public Implementation of ChIRo from "Learning 3D Representations of Molecular Chirality with Invariance to Bond Rotations"
Learning 3D Representations of Molecular Chirality with Invariance to Bond Rotations This directory contains the model architectures and experimental
Python Rapid Artificial Intelligence Ab Initio Molecular Dynamics
Python Rapid Artificial Intelligence Ab Initio Molecular Dynamics
3D-Transformer: Molecular Representation with Transformer in 3D Space
3D-Transformer: Molecular Representation with Transformer in 3D Space
Automatic Differentiation Multipole Moment Molecular Forcefield
Automatic Differentiation Multipole Moment Molecular Forcefield Performance notes On a single gpu, using waterbox_31ang.pdb example from MPIDplugin wh
Molecular AutoEncoder in PyTorch
MolEncoder Molecular AutoEncoder in PyTorch Install $ git clone https://github.com/cxhernandez/molencoder.git && cd molencoder $ python setup.py insta

DockStream: A Docking Wrapper to Enhance De Novo Molecular Design
DockStream Description DockStream is a docking wrapper providing access to a collection of ligand embedders and docking backends. Docking execution an

Code for the paper "JANUS: Parallel Tempered Genetic Algorithm Guided by Deep Neural Networks for Inverse Molecular Design"
JANUS: Parallel Tempered Genetic Algorithm Guided by Deep Neural Networks for Inverse Molecular Design This repository contains code for the paper: JA

Fast and scalable uncertainty quantification for neural molecular property prediction, accelerated optimization, and guided virtual screening.
Evidential Deep Learning for Guided Molecular Property Prediction and Discovery Ava Soleimany*, Alexander Amini*, Samuel Goldman*, Daniela Rus, Sangee
source code for https://arxiv.org/abs/2005.11248 "Accelerating Antimicrobial Discovery with Controllable Deep Generative Models and Molecular Dynamics"
Accelerating Antimicrobial Discovery with Controllable Deep Generative Models and Molecular Dynamics This work will be published in Nature Biomedical

Squidpy is a tool for the analysis and visualization of spatial molecular data.
Squidpy is a tool for the analysis and visualization of spatial molecular data. It builds on top of scanpy and anndata, from which it inherits modularity and scalability. It provides analysis tools that leverages the spatial coordinates of the data, as well as tissue images if available.
Kaggle | 9th place (part of) solution for the Bristol-Myers Squibb – Molecular Translation challenge
Part of the 9th place solution for the Bristol-Myers Squibb – Molecular Translation challenge translating images containing chemical structures into I

Implementation of Learning Gradient Fields for Molecular Conformation Generation (ICML 2021).
[PDF] | [Slides] The official implementation of Learning Gradient Fields for Molecular Conformation Generation (ICML 2021 Long talk) Installation Inst

MolRep: A Deep Representation Learning Library for Molecular Property Prediction
MolRep: A Deep Representation Learning Library for Molecular Property Prediction Summary MolRep is a Python package for fairly measuring algorithmic p
MDAnalysis tool to calculate membrane curvature.
The MDAkit for membrane curvature analysis is part of the Google Summer of Code program and it is linked to a Code of Conduct.
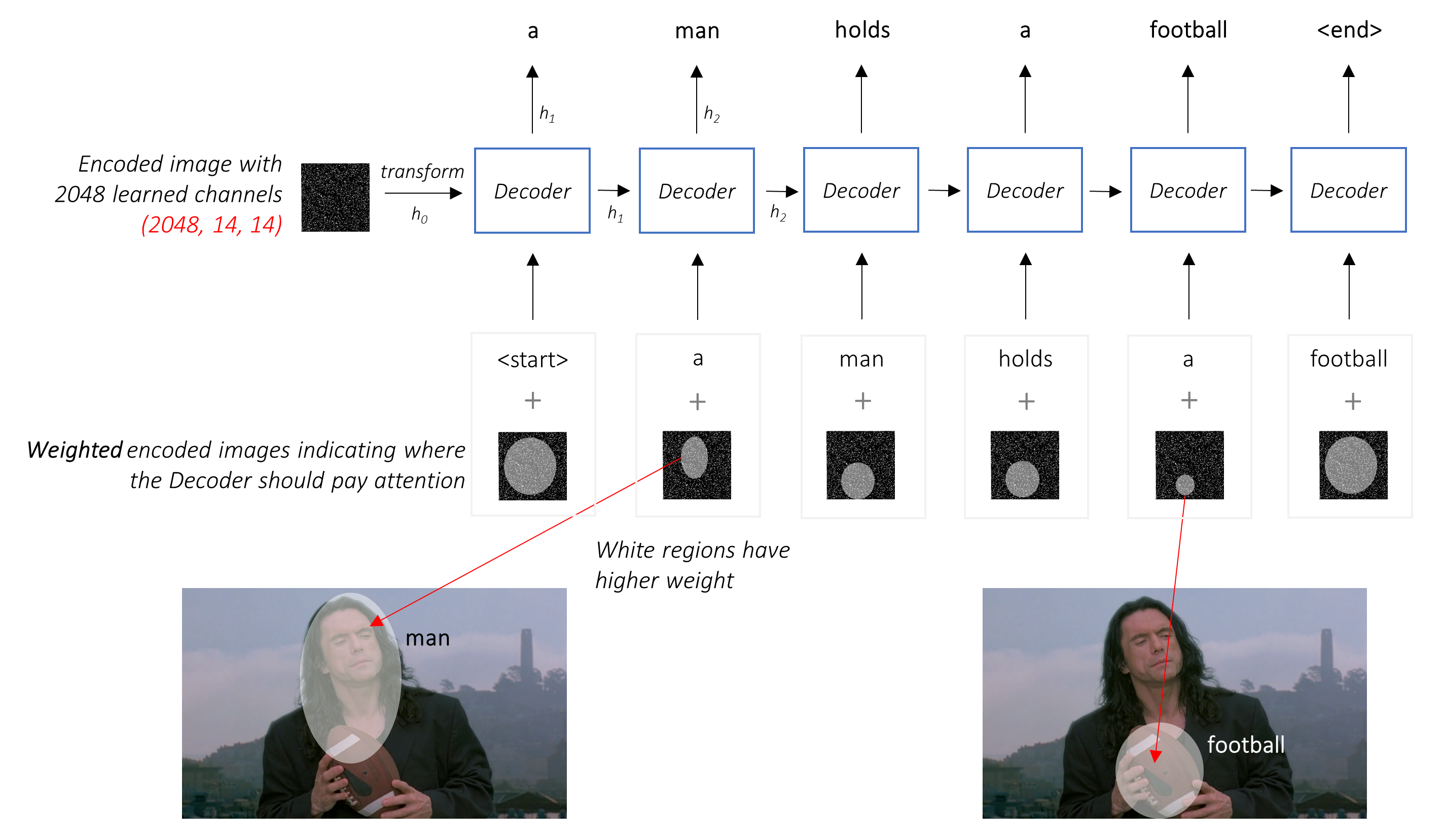
Pipeline for chemical image-to-text competition
BMS-Molecular-Translation Introduction This is a pipeline for Bristol-Myers Squibb – Molecular Translation by Vadim Timakin and Maksim Zhdanov. We got

SkipGNN: Predicting Molecular Interactions with Skip-Graph Networks (Scientific Reports)
SkipGNN: Predicting Molecular Interactions with Skip-Graph Networks Molecular interaction networks are powerful resources for the discovery. While dee
Few-Shot Graph Learning for Molecular Property Prediction
Few-shot Graph Learning for Molecular Property Prediction Introduction This is the source code and dataset for the following paper: Few-shot Graph Lea
Differentiable molecular simulation of proteins with a coarse-grained potential
Differentiable molecular simulation of proteins with a coarse-grained potential This repository contains the learned potential, simulation scripts and
This is a Pytorch implementation of the paper: Self-Supervised Graph Transformer on Large-Scale Molecular Data.
This is a Pytorch implementation of the paper: Self-Supervised Graph Transformer on Large-Scale Molecular Data.