58 Repositories
Python cell-biology Libraries

Jupyter Dock is a set of Jupyter Notebooks for performing molecular docking protocols interactively, as well as visualizing, converting file formats and analyzing the results.
Molecular Docking integrated in Jupyter Notebooks Description | Citation | Installation | Examples | Limitations | License Table of content Descriptio

TigerLily: Finding drug interactions in silico with the Graph.
Drug Interaction Prediction with Tigerlily Documentation | Example Notebook | Youtube Video | Project Report Tigerlily is a TigerGraph based system de

Implementation of GeoDiff: a Geometric Diffusion Model for Molecular Conformation Generation (ICLR 2022).
GeoDiff: a Geometric Diffusion Model for Molecular Conformation Generation [OpenReview] [arXiv] [Code] The official implementation of GeoDiff: A Geome

MarcoPolo is a clustering-free approach to the exploration of bimodally expressed genes along with group information in single-cell RNA-seq data
MarcoPolo is a method to discover differentially expressed genes in single-cell RNA-seq data without depending on prior clustering Overview MarcoPolo

Addon and nodes for working with structural biology and molecular data in Blender.
Molecular Nodes 🧬 🔬 💻 Buy Me a Coffee to Keep Development Going! Join a Community of Blender SciVis People! What is Molecular Nodes? Molecular Node

scAR (single-cell Ambient Remover) is a package for data denoising in single-cell omics.
scAR scAR (single cell Ambient Remover) is a package for denoising multiple single cell omics data. It can be used for multiple tasks, such as, sgRNA

Image-to-image regression with uncertainty quantification in PyTorch
Image-to-image regression with uncertainty quantification in PyTorch. Take any dataset and train a model to regress images to images with rigorous, distribution-free uncertainty quantification.
SBINN: Systems-biology informed neural network
SBINN: Systems-biology informed neural network The source code for the paper M. Daneker, Z. Zhang, G. E. Karniadakis, & L. Lu. Systems biology: Identi

Colab-xterm allows you to open a terminal in a cell
colab-xterm Colab-xterm allows you to open a terminal in a cell. Usage Install package and load the extension !pip install git+https://github.com/popc

Reverse engineering the dengue virus (under development construction)
Reverse engineering the dengue virus (under development 🚧 ) What is dengue? Dengue is a viral infection transmitted to humans through the bite of inf
Official repository for the ISBI 2021 paper Transformer Assisted Convolutional Neural Network for Cell Instance Segmentation
SegPC-2021 This is the official repository for the ISBI 2021 paper Transformer Assisted Convolutional Neural Network for Cell Instance Segmentation by
The repository includes the code for training cell counting applications. (Keras + Tensorflow)
cell_counting_v2 The repository includes the code for training cell counting applications. (Keras + Tensorflow) Dataset can be downloaded here : http:
BioPy is a collection (in-progress) of biologically-inspired algorithms written in Python
BioPy is a collection (in-progress) of biologically-inspired algorithms written in Python. Some of the algorithms included are mor
Single Red Blood Cell Hydrodynamic Traps Via the Generative Design
Rbc-traps-generative-design - The generative design for single red clood cell hydrodynamic traps using GEFEST framework
CellRank's reproducibility repository.
CellRank's reproducibility repository We believe that reproducibility is key and have made it as simple as possible to reproduce our results. Please e
Solution of Kaggle competition: Sartorius - Cell Instance Segmentation
Sartorius - Cell Instance Segmentation https://www.kaggle.com/c/sartorius-cell-instance-segmentation Environment setup Build docker image bash .dev_sc
Excel cell checker with python
excel-cell-checker Description This tool checks a given .xlsx file has the struc

Spatial Single-Cell Analysis Toolkit
Single-Cell Image Analysis Package Scimap is a scalable toolkit for analyzing spatial molecular data. The underlying framework is generalizable to spa
NeoDTI: Neural integration of neighbor information from a heterogeneous network for discovering new drug-target interactions
NeoDTI NeoDTI: Neural integration of neighbor information from a heterogeneous network for discovering new drug-target interactions (Bioinformatics).
A pythonic interface to high-throughput virtual screening software
pyscreener A pythonic interface to high-throughput virtual screening software Overview This repository contains the source of pyscreener, both a libra

A Parameter-free Deep Embedded Clustering Method for Single-cell RNA-seq Data
A Parameter-free Deep Embedded Clustering Method for Single-cell RNA-seq Data Overview Clustering analysis is widely utilized in single-cell RNA-seque

A PyTorch based deep learning library for drug pair scoring.
Documentation | External Resources | Datasets | Examples ChemicalX is a deep learning library for drug-drug interaction, polypharmacy side effect and

Full Spectrum Bioinformatics - a free online text designed to introduce key topics in Bioinformatics using the Python
Full Spectrum Bioinformatics is a free online text designed to introduce key topics in Bioinformatics using the Python programming language. The text is written in interactive Jupyter Notebooks, which allow you to try out and modify example code and analyses.

Python script to generate vector graphics of an oriented lattice unit cell
unitcell Python script to generate vector graphics of an oriented lattice unit cell Examples unitcell --type hexagonal --eulers 12 23 34 --axes --crys

Functions to analyze Cell-ID single-cell cytometry data using python language.
PyCellID (building...) Functions to analyze Cell-ID single-cell cytometry data using python language. Dependecies for this project. attrs(=21.1.0) fo

Implementation of the ivis algorithm as described in the paper Structure-preserving visualisation of high dimensional single-cell datasets.
Implementation of the ivis algorithm as described in the paper Structure-preserving visualisation of high dimensional single-cell datasets.

Codes for our paper The Stem Cell Hypothesis: Dilemma behind Multi-Task Learning with Transformer Encoders published to EMNLP 2021.
The Stem Cell Hypothesis Codes for our paper The Stem Cell Hypothesis: Dilemma behind Multi-Task Learning with Transformer Encoders published to EMNLP
Single-Cell Analysis in Python. Scales to 1M cells.
Scanpy – Single-Cell Analysis in Python Scanpy is a scalable toolkit for analyzing single-cell gene expression data built jointly with anndata. It inc

MICOM is a Python package for metabolic modeling of microbial communities
Welcome MICOM is a Python package for metabolic modeling of microbial communities currently developed in the Gibbons Lab at the Institute for Systems
Single cell current best practices tutorial case study for the paper:Luecken and Theis, "Current best practices in single-cell RNA-seq analysis: a tutorial"
Scripts for "Current best-practices in single-cell RNA-seq: a tutorial" This repository is complementary to the publication: M.D. Luecken, F.J. Theis,

A generalist algorithm for cell and nucleus segmentation.
Cellpose | A generalist algorithm for cell and nucleus segmentation. Cellpose was written by Carsen Stringer and Marius Pachitariu. To learn about Cel
t-SNE and hierarchical clustering are popular methods of exploratory data analysis, particularly in biology.
tree-SNE t-SNE and hierarchical clustering are popular methods of exploratory data analysis, particularly in biology. Building on recent advances in s
Subpopulation detection in high-dimensional single-cell data
PhenoGraph for Python3 PhenoGraph is a clustering method designed for high-dimensional single-cell data. It works by creating a graph ("network") repr
A Python adaption of Augur to prioritize cell types in perturbation analysis.
A Python adaption of Augur to prioritize cell types in perturbation analysis.

A generalist algorithm for cell and nucleus segmentation.
Cellpose | A generalist algorithm for cell and nucleus segmentation. Cellpose was written by Carsen Stringer and Marius Pachitariu. To learn about Cel
A tool to visualise the results of AlphaFold2 and inspect the quality of structural predictions
AlphaFold Analyser This program produces high quality visualisations of predicted structures produced by AlphaFold. These visualisations allow the use
Michael Vinyard's utilities
Install vintools To download this package from pypi: pip install vintools Install the development package To download and install the developmen

Map single-cell transcriptomes to copy number evolutionary trees.
Map single-cell transcriptomes to copy number evolutionary trees. Check out the tutorial for more information. Installation $ pip install scatrex SCA
ToeholdTools is a Python package and desktop app designed to facilitate analyzing and designing toehold switches, created as part of the 2021 iGEM competition.
ToeholdTools Category Status Repository Package Build Quality A library for the analysis of toehold switch riboregulators created by the iGEM team Cit

A customized interface for single cell track visualisation based on pcnaDeep and napari.
pcnaDeep-napari A customized interface for single cell track visualisation based on pcnaDeep and napari. 👀 Under construction You can get test image

Official code for On Path Integration of Grid Cells: Group Representation and Isotropic Scaling (NeurIPS 2021)
On Path Integration of Grid Cells: Group Representation and Isotropic Scaling This repo contains the official implementation for the paper On Path Int
A totally unrealistic cell growth/reproduction simulation.
A totally unrealistic cell growth/reproduction simulation.

Kaggle: Cell Instance Segmentation
Kaggle: Cell Instance Segmentation The goal of this challenge is to detect cells in microscope images. with simple view on how many cels have been ann

A lightweight Python-based 3D network multi-agent simulator. Uses a cell-based congestion model. Calculates risk, loudness and battery capacities of the agents. Suitable for 3D network optimization tasks.
AMAZ3DSim AMAZ3DSim is a lightweight python-based 3D network multi-agent simulator. It uses a cell-based congestion model. It calculates risk, battery

starfish is a Python library for processing images of image-based spatial transcriptomics.
starfish: scalable pipelines for image-based transcriptomics starfish is a Python library for processing images of image-based spatial transcriptomics

BioMASS - A Python Framework for Modeling and Analysis of Signaling Systems
Mathematical modeling is a powerful method for the analysis of complex biological systems. Although there are many researches devoted on produ

Biomarker identification for COVID-19 Severity in BALF cells Single-cell RNA-seq data
scBALF Covid-19 dataset Analysis Here is the Github page that has the codes for the bioinformatics pipeline described in the paper COVID-Datathon: Bio
LIVECell - A large-scale dataset for label-free live cell segmentation
LIVECell dataset This document contains instructions of how to access the data associated with the submitted manuscript "LIVECell - A large-scale data

pcnaDeep integrates cutting-edge detection techniques with tracking and cell cycle resolving models.
pcnaDeep: a deep-learning based single-cell cycle profiler with PCNA signal Welcome! pcnaDeep integrates cutting-edge detection techniques with tracki
Main repository for the HackBio'2021 Virtual Internship Experience for #Team-Greider ❤️
Hello 🤟 #Team-Greider The team of 20 people for HackBio'2021 Virtual Bioinformatics Internship 💝 🖨️ 👨💻 HackBio: https://thehackbio.com 💬 Ask us

Message Passing on Cell Complexes
CW Networks This repository contains the code used for the papers Weisfeiler and Lehman Go Cellular: CW Networks (Under review) and Weisfeiler and Leh
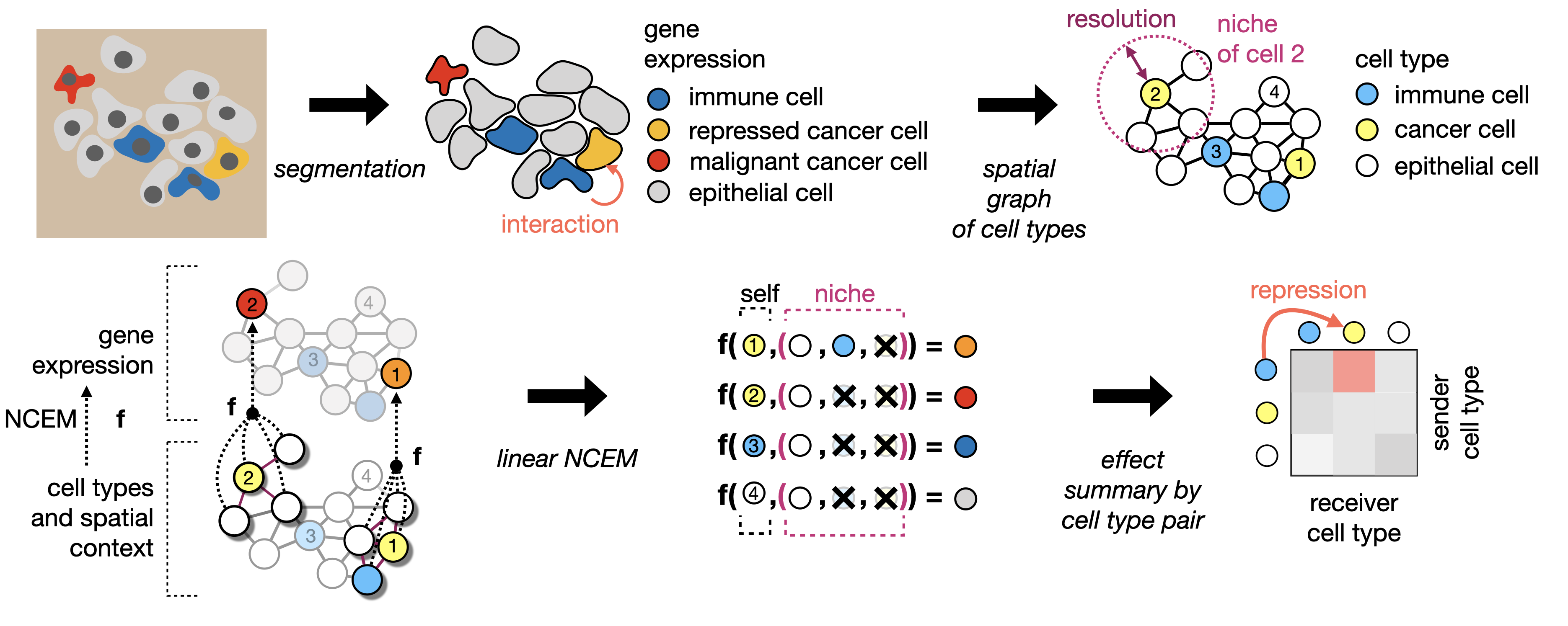
Learning cell communication from spatial graphs of cells
ncem Features Repository for the manuscript Fischer, D. S., Schaar, A. C. and Theis, F. Learning cell communication from spatial graphs of cells. 2021

A scanpy extension to analyse single-cell TCR and BCR data.
Scirpy: A Scanpy extension for analyzing single-cell immune-cell receptor sequencing data Scirpy is a scalable python-toolkit to analyse T cell recept
7th place solution of Human Protein Atlas - Single Cell Classification on Kaggle
kaggle-hpa-2021-7th-place-solution Code for 7th place solution of Human Protein Atlas - Single Cell Classification on Kaggle. A description of the met

Interpretation of T cell states using reference single-cell atlases
Interpretation of T cell states using reference single-cell atlases ProjecTILs is a computational method to project scRNA-seq data into reference sing

OPEM (Open Source PEM Fuel Cell Simulation Tool)
Table of contents What is PEM? Overview Installation Usage Executable Library Telegram Bot Try OPEM in Your Browser! MATLAB Issues & Bug Reports Contr

An interactive explorer for single-cell transcriptomics data
an interactive explorer for single-cell transcriptomics data cellxgene (pronounced "cell-by-gene") is an interactive data explorer for single-cell tra
Inner ear models for Python
cochlea cochlea is a collection of inner ear models. All models are easily accessible as Python functions. They take sound signal as input and return