106 Repositories
Python protein-sequences Libraries

SARS-Cov-2 Recombinant Finder for fasta sequences
Sc2rf - SARS-Cov-2 Recombinant Finder Pronounced: Scarf What's this? Sc2rf can search genome sequences of SARS-CoV-2 for potential recombinants - new

Jupyter Dock is a set of Jupyter Notebooks for performing molecular docking protocols interactively, as well as visualizing, converting file formats and analyzing the results.
Molecular Docking integrated in Jupyter Notebooks Description | Citation | Installation | Examples | Limitations | License Table of content Descriptio

RITA is a family of autoregressive protein models, developed by LightOn in collaboration with the OATML group at Oxford and the Debora Marks Lab at Harvard.
RITA: a Study on Scaling Up Generative Protein Sequence Models RITA is a family of autoregressive protein models, developed by a collaboration of Ligh
A library built upon PyTorch for building embeddings on discrete event sequences using self-supervision
pytorch-lifestream a library built upon PyTorch for building embeddings on discrete event sequences using self-supervision. It can process terabyte-si
Contains the code and data for our #ICSE2022 paper titled as "CodeFill: Multi-token Code Completion by Jointly Learning from Structure and Naming Sequences"
CodeFill This repository contains the code for our paper titled as "CodeFill: Multi-token Code Completion by Jointly Learning from Structure and Namin

Persian Bert For Long-Range Sequences
ParsBigBird: Persian Bert For Long-Range Sequences The Bert and ParsBert algorithms can handle texts with token lengths of up to 512, however, many ta

Implementation of a protein autoregressive language model, but with autoregressive infilling objective (editing subsequences capability)
Protein GLM (wip) Implementation of a protein autoregressive language model, but with autoregressive infilling objective (editing subsequences capabil

Implementation of the GVP-Transformer, which was used in the paper "Learning inverse folding from millions of predicted structures" for de novo protein design alongside Alphafold2
GVP Transformer (wip) Implementation of the GVP-Transformer, which was used in the paper Learning inverse folding from millions of predicted structure

Official implementation of "Generating 3D Molecules for Target Protein Binding"
Generating 3D Molecules for Target Protein Binding This is the official implementation of the GraphBP method proposed in the following paper. Meng Liu

Addon and nodes for working with structural biology and molecular data in Blender.
Molecular Nodes 🧬 🔬 💻 Buy Me a Coffee to Keep Development Going! Join a Community of Blender SciVis People! What is Molecular Nodes? Molecular Node

ProtFeat is protein feature extraction tool that utilizes POSSUM and iFeature.
Description: ProtFeat is designed to extract the protein features by employing POSSUM and iFeature python-based tools. ProtFeat includes a total of 39
Tuple-sum-filter - Library to play with filtering numeric sequences by sums of their pairs, triplets, etc. With a bonus CLI demo
Tuple Sum Filter A library to play with filtering numeric sequences by sums of t

Neighbor2Seq: Deep Learning on Massive Graphs by Transforming Neighbors to Sequences
Neighbor2Seq: Deep Learning on Massive Graphs by Transforming Neighbors to Sequences This repository is an official PyTorch implementation of Neighbor

EquiBind: Geometric Deep Learning for Drug Binding Structure Prediction
EquiBind: geometric deep learning for fast predictions of the 3D structure in which a small molecule binds to a protein

Benchmarking Pipeline for Prediction of Protein-Protein Interactions
B4PPI Benchmarking Pipeline for the Prediction of Protein-Protein Interactions How this benchmarking pipeline has been built, and how to use it, is de
A small tool to test and visualize protein embeddings and amino acid proportions.
polyprotein_stats A small tool to test and visualize protein embeddings and amino acid proportions. Currently deployed on streamlit.io. Given a set of
Doughskript interpreter for converting simple command sequences into executable Arduino C++ code.
Doughskript interpreter for converting simple command sequences into executable Arduino C++ code.

This repository contains the implementation of the paper Contrastive Instance Association for 4D Panoptic Segmentation using Sequences of 3D LiDAR Scans
Contrastive Instance Association for 4D Panoptic Segmentation using Sequences of 3D LiDAR Scans This repository contains the implementation of the pap

Source code for EquiDock: Independent SE(3)-Equivariant Models for End-to-End Rigid Protein Docking (ICLR 2022)
Source code for EquiDock: Independent SE(3)-Equivariant Models for End-to-End Rigid Protein Docking (ICLR 2022) Please cite "Independent SE(3)-Equivar
Codes and models for the paper "Learning Unknown from Correlations: Graph Neural Network for Inter-novel-protein Interaction Prediction".
GNN_PPI Codes and models for the paper "Learning Unknown from Correlations: Graph Neural Network for Inter-novel-protein Interaction Prediction". Lear

OntoProtein: Protein Pretraining With Ontology Embedding
OntoProtein This is the implement of the paper "OntoProtein: Protein Pretraining With Ontology Embedding". OntoProtein is an effective method that mak
Improved Fitness Optimization Landscapes for Sequence Design
ReLSO Improved Fitness Optimization Landscapes for Sequence Design Description Citation How to run Training models Original data source Description In
Ingestinator is my personal VFX pipeline tool for ingesting folders containing frame sequences that have been pulled and downloaded to a local folder
Ingestinator Ingestinator is my personal VFX pipeline tool for ingesting folders containing frame sequences that have been pulled and downloaded to a

Linux GUI app to codon optimize many single-fasta files with coding sequences , using many taxonomy ids
codon_optimize_cds_with_many_taxids_singlefasta Linux GUI app to codon optimize many single-fasta files with coding sequences, using many taxonomy ids
PROTEIN EXPRESSION ANALYSIS FOR DOWN SYNDROME
PROTEIN-EXPRESSION-ANALYSIS-FOR-DOWN-SYNDROME Down syndrome (DS) is a chromosomal disorder where organisms have an extra chromosome 21, sometimes know

'rl_UK' is an open-source command-line tool in Python for calculating the shortest path between BUS stop sequences in the UK
'rl_UK' is an open-source command-line tool in Python for calculating the shortest path between BUS stop sequences in the UK. As input files, it uses an ATCO-CIF file and 'OS Open Roads' dataset from Ordnance Survey Data Hub.

Deep learning transformer model that generates unique music sequences.
music-ai Deep learning transformer model that generates unique music sequences. Abstract In 2017, a new state-of-the-art was published for natural lan

Implementation of Memory-Compressed Attention, from the paper "Generating Wikipedia By Summarizing Long Sequences"
Memory Compressed Attention Implementation of the Self-Attention layer of the proposed Memory-Compressed Attention, in Pytorch. This repository offers
Continual Learning of Long Topic Sequences in Neural Information Retrieval
ContinualPassageRanking Repository for the paper "Continual Learning of Long Topic Sequences in Neural Information Retrieval". In this repository you
RFDesign - Protein hallucination and inpainting with RoseTTAFold
RFDesign: Protein hallucination and inpainting with RoseTTAFold Jue Wang (juewan
The official code of "SCROLLS: Standardized CompaRison Over Long Language Sequences".
SCROLLS This repository contains the official code of the paper: "SCROLLS: Standardized CompaRison Over Long Language Sequences". Links Official Websi
LSTM built using Keras Python package to predict time series steps and sequences. Includes sin wave and stock market data
LSTM Neural Network for Time Series Prediction LSTM built using the Keras Python package to predict time series steps and sequences. Includes sine wav

Official implementation for the paper: Generating Smooth Pose Sequences for Diverse Human Motion Prediction
Generating Smooth Pose Sequences for Diverse Human Motion Prediction This is official implementation for the paper Generating Smooth Pose Sequences fo
A fast Protein Chain / Ligand Extractor and organizer.
Are you tired of using visualization software, or full blown suites just to separate protein chains / ligands ? Are you tired of organizing the mess o
This program analyzes a DNA sequence and outputs snippets of DNA that are likely to be protein-coding genes.
This program analyzes a DNA sequence and outputs snippets of DNA that are likely to be protein-coding genes.

Retrieve annotated intron sequences and classify them as minor (U12-type) or major (U2-type)
(intron I nterrogator and C lassifier) intronIC is a program that can be used to classify intron sequences as minor (U12-type) or major (U2-type), usi
Annotates sequences with Eggnog-mapper and hhblits against PDB70
Annotating "hypothetical" proteins with the PDB See config/ for configuration information. This workflow takes as input a set of protein sequences. It
SeqLike - flexible biological sequence objects in Python
SeqLike - flexible biological sequence objects in Python Introduction A single object API that makes working with biological sequences in Python more
Examples of using sparse attention, as in "Generating Long Sequences with Sparse Transformers"
Status: Archive (code is provided as-is, no updates expected) Update August 2020: For an example repository that achieves state-of-the-art modeling pe

Infrastructure template and Jupyter notebooks for running RoseTTAFold on AWS Batch.
AWS RoseTTAFold Infrastructure template and Jupyter notebooks for running RoseTTAFold on AWS Batch. Overview Proteins are large biomolecules that play

GebPy is a Python-based, open source tool for the generation of geological data of minerals, rocks and complete lithological sequences.
GebPy is a Python-based, open source tool for the generation of geological data of minerals, rocks and complete lithological sequences. The data can be generated randomly or with respect to user-defined constraints, for example a specific element concentration within minerals and rocks or the order of units within a complete lithological profile.

A minimal, standalone viewer for 3D animations stored as stop-motion sequences of individual .obj mesh files.
ObjSequenceViewer V0.5 A minimal, standalone viewer for 3D animations stored as stop-motion sequences of individual .obj mesh files. Installation: pip
Codes for TIM2021 paper "Anchor-Based Spatio-Temporal Attention 3-D Convolutional Networks for Dynamic 3-D Point Cloud Sequences"
Codes for TIM2021 paper "Anchor-Based Spatio-Temporal Attention 3-D Convolutional Networks for Dynamic 3-D Point Cloud Sequences"
![[NeurIPS 2021] Garment4D: Garment Reconstruction from Point Cloud Sequences](https://github.com/hongfz16/Garment4D/raw/main/assets/cover.jpg)
[NeurIPS 2021] Garment4D: Garment Reconstruction from Point Cloud Sequences
Garment4D [PDF] | [OpenReview] | [Project Page] Overview This is the codebase for our NeurIPS 2021 paper Garment4D: Garment Reconstruction from Point
Uni-Fold: Training your own deep protein-folding models.
Uni-Fold: Training your own deep protein-folding models. This package provides and implementation of a trainable, Transformer-based deep protein foldi

This is an official PyTorch implementation of "Contrastive Learning from Extremely Augmented Skeleton Sequences for Self-supervised Action Recognition" in AAAI2022.
AimCLR This is an official PyTorch implementation of "Contrastive Learning from Extremely Augmented Skeleton Sequences for Self-supervised Action Reco
![[NeurIPS 2021] Garment4D: Garment Reconstruction from Point Cloud Sequences](https://github.com/hongfz16/Garment4D/raw/main/assets/cover.jpg)
[NeurIPS 2021] Garment4D: Garment Reconstruction from Point Cloud Sequences
Garment4D [PDF] | [OpenReview] | [Project Page] Overview This is the codebase for our NeurIPS 2021 paper Garment4D: Garment Reconstruction from Point
![[NeurIPS 2021] COCO-LM: Correcting and Contrasting Text Sequences for Language Model Pretraining](https://github.com/microsoft/COCO-LM/raw/main/coco-lm.png)
[NeurIPS 2021] COCO-LM: Correcting and Contrasting Text Sequences for Language Model Pretraining
COCO-LM This repository contains the scripts for fine-tuning COCO-LM pretrained models on GLUE and SQuAD 2.0 benchmarks. Paper: COCO-LM: Correcting an
Uni-Fold: Training your own deep protein-folding models
Uni-Fold: Training your own deep protein-folding models. This package provides an implementation of a trainable, Transformer-based deep protein foldin

Official PyTorch implementation of "Contrastive Learning from Extremely Augmented Skeleton Sequences for Self-supervised Action Recognition" in AAAI2022.
AimCLR This is an official PyTorch implementation of "Contrastive Learning from Extremely Augmented Skeleton Sequences for Self-supervised Action Reco
Using deep learning to predict gene structures of the coding genes in DNA sequences of Arabidopsis thaliana
DeepGeneAnnotator: A tool to annotate the gene in the genome The master thesis of the "Using deep learning to predict gene structures of the coding ge

Decoding the Protein-ligand Interactions Using Parallel Graph Neural Networks
Decoding the Protein-ligand Interactions Using Parallel Graph Neural Networks Requirements python 0.10+ rdkit 2020.03.3.0 biopython 1.78 openbabel 2.4

Tools to create pixel-wise object masks, bounding box labels (2D and 3D) and 3D object model (PLY triangle mesh) for object sequences filmed with an RGB-D camera.
Tools to create pixel-wise object masks, bounding box labels (2D and 3D) and 3D object model (PLY triangle mesh) for object sequences filmed with an RGB-D camera. This project prepares training and testing data for various deep learning projects such as 6D object pose estimation projects singleshotpose, as well as object detection and instance segmentation projects.
Sequence clustering and database creation using mmseqs, from local fasta files
Sequence clustering and database creation using mmseqs, from local fasta files

A Protein-RNA Interface Predictor Based on Semantics of Sequences
PRIP PRIP:A Protein-RNA Interface Predictor Based on Semantics of Sequences installation gensim==3.8.3 matplotlib==3.1.3 xgboost==1.3.3 prettytable==2
A package to predict protein inter-residue geometries from sequence data
trRosetta This package is a part of trRosetta protein structure prediction protocol developed in: Improved protein structure prediction using predicte

Bio-Computing Platform Featuring Large-Scale Representation Learning and Multi-Task Deep Learning “螺旋桨”生物计算工具集
English | 简体中文 Latest News 2021.10.25 Paper "Docking-based Virtual Screening with Multi-Task Learning" is accepted by BIBM 2021. 2021.07.29 PaddleHeli
ARAE-Tensorflow for Discrete Sequences (Adversarially Regularized Autoencoder)
ARAE Tensorflow Code Code for the paper Adversarially Regularized Autoencoders for Generating Discrete Structures by Zhao, Kim, Zhang, Rush and LeCun

MASS (Mueen's Algorithm for Similarity Search) - a python 2 and 3 compatible library used for searching time series sub-sequences under z-normalized Euclidean distance for similarity.
Introduction MASS allows you to search a time series for a subquery resulting in an array of distances. These array of distances enable you to identif

Addon for adding subtitle files to blender VSE as Text sequences. Using pysub2 python module.
Import Subtitles for Blender VSE Addon for adding subtitle files to blender VSE as Text sequences. Using pysub2 python module. Supported formats by py
A pipeline that creates consensus sequences from a Nanopore reads. I
A pipeline that creates consensus sequences from a Nanopore reads. It clusters reads that are similar to each other and creates a consensus that is then identified using BLAST.

This package is a python library with tools for the Molecular Simulation - Software Gromos.
This package is a python library with tools for the Molecular Simulation - Software Gromos. It allows you to easily set up, manage and analyze simulations in python.
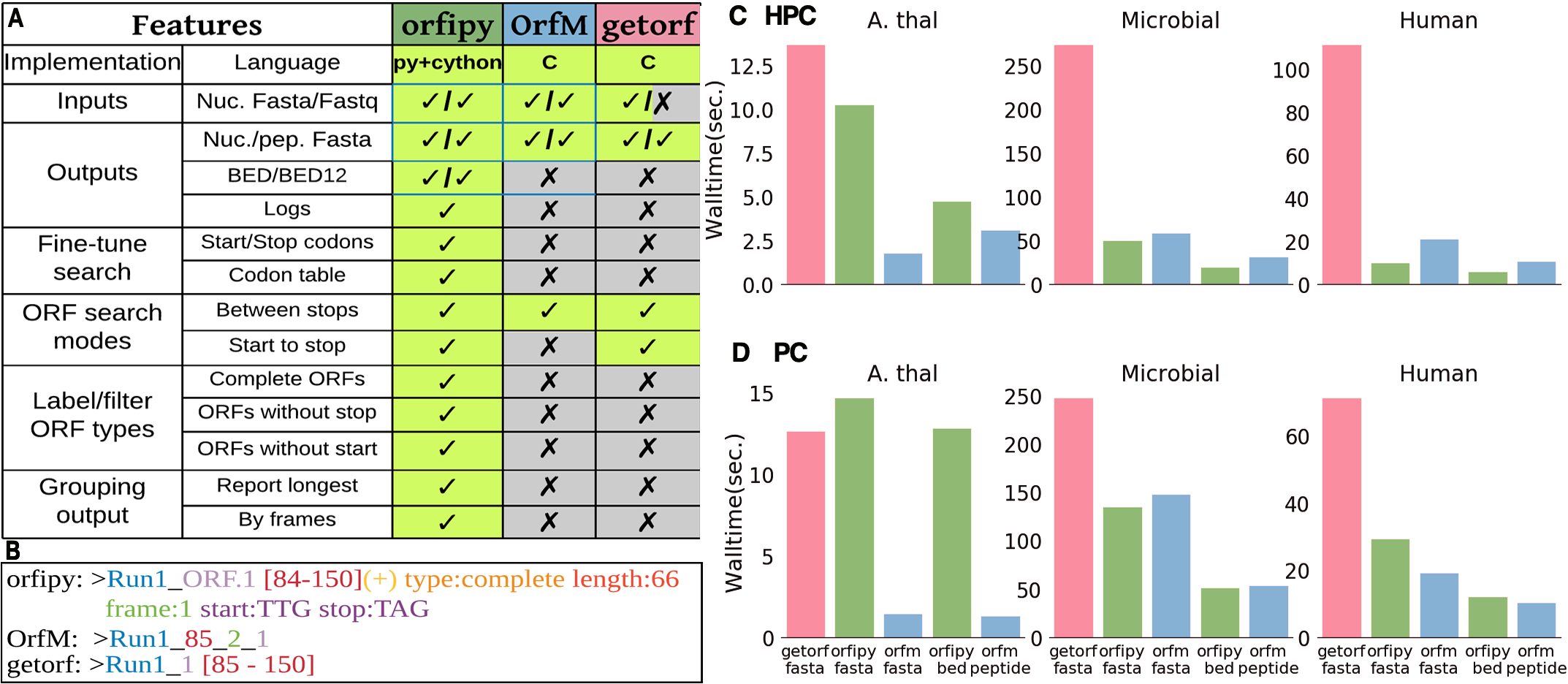
orfipy is a tool written in python/cython to extract ORFs in an extremely and fast and flexible manner
Introduction orfipy is a tool written in python/cython to extract ORFs in an extremely and fast and flexible manner. Other popular ORF searching tools

Bioinformatics tool for exploring RNA-Protein interactions
Explore RNA-Protein interactions. RNPFind is a bioinformatics tool. It takes an RNA transcript as input and gives a list of RNA binding protein (RBP)
Script For Importing Image sequences into scrap mechanic via blueprints
To use dowload and extract "video makes.zip" Python has to be installed https://www.python.org/ (may not work on version lower than 3.9) Has to be run
Deep generative models of 3D grids for structure-based drug discovery
What is liGAN? liGAN is a research codebase for training and evaluating deep generative models for de novo drug design based on 3D atomic density grid
Predicting lncRNA–protein interactions based on graph autoencoders and collaborative training
Predicting lncRNA–protein interactions based on graph autoencoders and collaborative training Code for our paper "Predicting lncRNA–protein interactio

Implementation of Enformer, Deepmind's attention network for predicting gene expression, in Pytorch
Enformer - Pytorch (wip) Implementation of Enformer, Deepmind's attention network for predicting gene expression, in Pytorch. The original tensorflow
Physicochemical properties and indices for amino-acid sequences (ported from R).
peptides.py Physicochemical properties and indices for amino-acid sequences. 🗺️ Overview peptides.py is a pure-Python package to compute common descr
peptides.py is a pure-Python package to compute common descriptors for protein sequences
peptides.py Physicochemical properties and indices for amino-acid sequences. 🗺️ Overview peptides.py is a pure-Python package to compute common descr
A graph neural network (GNN) model to predict protein-protein interactions (PPI) with no sample features
A graph neural network (GNN) model to predict protein-protein interactions (PPI) with no sample features
A geometric deep learning pipeline for predicting protein interface contacts.
A geometric deep learning pipeline for predicting protein interface contacts.
Fuzzy string matching like a boss. It uses Levenshtein Distance to calculate the differences between sequences in a simple-to-use package.
Fuzzy string matching like a boss. It uses Levenshtein Distance to calculate the differences between sequences in a simple-to-use package.
Beyond Paragraphs: NLP for Long Sequences
Beyond Paragraphs: NLP for Long Sequences

Sign Language is detected in realtime using video sequences. Our approach involves MediaPipe Holistic for keypoints extraction and LSTM Model for prediction.
RealTime Sign Language Detection using Action Recognition Approach Real-Time Sign Language is commonly predicted using models whose architecture consi

Implementation of Neural Distance Embeddings for Biological Sequences (NeuroSEED) in PyTorch
Neural Distance Embeddings for Biological Sequences Official implementation of Neural Distance Embeddings for Biological Sequences (NeuroSEED) in PyTo
Python library for generating sequences with uniform stimulus history
Sampling Euler tours for uniform stimulus history Table of Contents About Examples Experiment 1 Experiment 2 Experiment 3 Experiment 4 Experiment 5 Co
Replication attempt for the Protein Folding Model
RGN2-Replica (WIP) To eventually become an unofficial working Pytorch implementation of RGN2, an state of the art model for MSA-less Protein Folding f

Graph-based community clustering approach to extract protein domains from a predicted aligned error matrix
Using a predicted aligned error matrix corresponding to an AlphaFold2 model , returns a series of lists of residue indices, where each list corresponds to a set of residues clustering together into a pseudo-rigid domain.

A simple yet powerful TUI framework for your Python (3.7+) applications
A simple yet powerful TUI framework for your Python (3.7+) applications

Implementation of the Triangle Multiplicative module, used in Alphafold2 as an efficient way to mix rows or columns of a 2d feature map, as a standalone package for Pytorch
Triangle Multiplicative Module - Pytorch Implementation of the Triangle Multiplicative module, used in Alphafold2 as an efficient way to mix rows or c

Implementation of Invariant Point Attention, used for coordinate refinement in the structure module of Alphafold2, as a standalone Pytorch module
Invariant Point Attention - Pytorch Implementation of Invariant Point Attention as a standalone module, which was used in the structure module of Alph
Implementation and replication of ProGen, Language Modeling for Protein Generation, in Jax
ProGen - (wip) Implementation and replication of ProGen, Language Modeling for Protein Generation, in Pytorch and Jax (the weights will be made easily
7th place solution of Human Protein Atlas - Single Cell Classification on Kaggle
kaggle-hpa-2021-7th-place-solution Code for 7th place solution of Human Protein Atlas - Single Cell Classification on Kaggle. A description of the met

A denoising diffusion probabilistic model (DDPM) tailored for conditional generation of protein distograms
Denoising Diffusion Probabilistic Model for Proteins Implementation of Denoising Diffusion Probabilistic Model in Pytorch. It is a new approach to gen

Unofficial TensorFlow implementation of Protein Interface Prediction using Graph Convolutional Networks.
[TensorFlow] Protein Interface Prediction using Graph Convolutional Networks Unofficial TensorFlow implementation of Protein Interface Prediction usin

Model-free Vehicle Tracking and State Estimation in Point Cloud Sequences
Model-free Vehicle Tracking and State Estimation in Point Cloud Sequences 1. Introduction This project is for paper Model-free Vehicle Tracking and St
Protein Language Model
ProteinLM We pretrain protein language model based on Megatron-LM framework, and then evaluate the pretrained model results on TAPE (Tasks Assessing P
Implementation of ProteinBERT in Pytorch
ProteinBERT - Pytorch (wip) Implementation of ProteinBERT in Pytorch. Original Repository Install $ pip install protein-bert-pytorch Usage import torc
Explore related sequences in the OEIS
OEIS explorer This is a tool for exploring two different kinds of relationships between sequences in the OEIS: mentions (links) of other sequences on
Implementation of ProteinBERT in Pytorch
ProteinBERT - Pytorch (wip) Implementation of ProteinBERT in Pytorch. Original Repository Install $ pip install protein-bert-pytorch Usage import torc

Official implementation of the network presented in the paper "M4Depth: A motion-based approach for monocular depth estimation on video sequences"
M4Depth This is the reference TensorFlow implementation for training and testing depth estimation models using the method described in M4Depth: A moti
ProteinBERT is a universal protein language model pretrained on ~106M proteins from the UniRef90 dataset.
ProteinBERT is a universal protein language model pretrained on ~106M proteins from the UniRef90 dataset. Through its Python API, the pretrained model can be fine-tuned on any protein-related task in a matter of minutes. Based on our experiments with a wide range of benchmarks, ProteinBERT usually achieves state-of-the-art performance. ProteinBERT is built on TenforFlow/Keras.

Deep functional residue identification
DeepFRI Deep functional residue identification Citing @article {Gligorijevic2019, author = {Gligorijevic, Vladimir and Renfrew, P. Douglas and Koscio

Generative Models for Graph-Based Protein Design
Graph-Based Protein Design This repo contains code for Generative Models for Graph-Based Protein Design by John Ingraham, Vikas Garg, Regina Barzilay
Structural basis for solubility in protein expression systems
Structural basis for solubility in protein expression systems Large-scale protein production for biotechnology and biopharmaceutical applications rely

Implementation of the "PSTNet: Point Spatio-Temporal Convolution on Point Cloud Sequences" paper.
PSTNet: Point Spatio-Temporal Convolution on Point Cloud Sequences Introduction Point cloud sequences are irregular and unordered in the spatial dimen

Implementation of COCO-LM, Correcting and Contrasting Text Sequences for Language Model Pretraining, in Pytorch
COCO LM Pretraining (wip) Implementation of COCO-LM, Correcting and Contrasting Text Sequences for Language Model Pretraining, in Pytorch. They were a
Differentiable molecular simulation of proteins with a coarse-grained potential
Differentiable molecular simulation of proteins with a coarse-grained potential This repository contains the learned potential, simulation scripts and

Compute distance between sequences. 30+ algorithms, pure python implementation, common interface, optional external libs usage.
TextDistance TextDistance -- python library for comparing distance between two or more sequences by many algorithms. Features: 30+ algorithms Pure pyt