191 Repositories
Python protein-structure Libraries

Jupyter Dock is a set of Jupyter Notebooks for performing molecular docking protocols interactively, as well as visualizing, converting file formats and analyzing the results.
Molecular Docking integrated in Jupyter Notebooks Description | Citation | Installation | Examples | Limitations | License Table of content Descriptio

Extracting Tables from Document Images using a Multi-stage Pipeline for Table Detection and Table Structure Recognition:
Multi-Type-TD-TSR Check it out on Source Code of our Paper: Multi-Type-TD-TSR Extracting Tables from Document Images using a Multi-stage Pipeline for
An excellent hash algorithm combining classical sponge structure and RNN.
SHA-RNN Recurrent Neural Network with Chaotic System for Hash Functions Anonymous Authors [摘要] 在这次作业中我们提出了一种新的 Hash Function —— SHA-RNN。其以海绵结构为基础,融合了混

RITA is a family of autoregressive protein models, developed by LightOn in collaboration with the OATML group at Oxford and the Debora Marks Lab at Harvard.
RITA: a Study on Scaling Up Generative Protein Sequence Models RITA is a family of autoregressive protein models, developed by a collaboration of Ligh

Incremental Transformer Structure Enhanced Image Inpainting with Masking Positional Encoding (CVPR2022)
Incremental Transformer Structure Enhanced Image Inpainting with Masking Positional Encoding by Qiaole Dong*, Chenjie Cao*, Yanwei Fu Paper and Supple
Contains the code and data for our #ICSE2022 paper titled as "CodeFill: Multi-token Code Completion by Jointly Learning from Structure and Naming Sequences"
CodeFill This repository contains the code for our paper titled as "CodeFill: Multi-token Code Completion by Jointly Learning from Structure and Namin
Make your first PR. A beginner friendly repository made specifically for open source beginners. Add any program under any language (it can be anything from a simple program to a complex data structure algorithm). Happy coding...
Hacktober Fest 2021 Upload Different Types of Programs in any Language Use this project to make your first contribution to an open source project on G

Implementation of a protein autoregressive language model, but with autoregressive infilling objective (editing subsequences capability)
Protein GLM (wip) Implementation of a protein autoregressive language model, but with autoregressive infilling objective (editing subsequences capabil

Implementation of the GVP-Transformer, which was used in the paper "Learning inverse folding from millions of predicted structures" for de novo protein design alongside Alphafold2
GVP Transformer (wip) Implementation of the GVP-Transformer, which was used in the paper Learning inverse folding from millions of predicted structure

Official implementation of "Generating 3D Molecules for Target Protein Binding"
Generating 3D Molecules for Target Protein Binding This is the official implementation of the GraphBP method proposed in the following paper. Meng Liu
Official codebase for ICLR oral paper Unsupervised Vision-Language Grammar Induction with Shared Structure Modeling
CLIORA This is the official codebase for ICLR oral paper: Unsupervised Vision-Language Grammar Induction with Shared Structure Modeling. We introduce

Addon and nodes for working with structural biology and molecular data in Blender.
Molecular Nodes 🧬 🔬 💻 Buy Me a Coffee to Keep Development Going! Join a Community of Blender SciVis People! What is Molecular Nodes? Molecular Node

ProtFeat is protein feature extraction tool that utilizes POSSUM and iFeature.
Description: ProtFeat is designed to extract the protein features by employing POSSUM and iFeature python-based tools. ProtFeat includes a total of 39

EquiBind: Geometric Deep Learning for Drug Binding Structure Prediction
EquiBind: geometric deep learning for fast predictions of the 3D structure in which a small molecule binds to a protein

Benchmarking Pipeline for Prediction of Protein-Protein Interactions
B4PPI Benchmarking Pipeline for the Prediction of Protein-Protein Interactions How this benchmarking pipeline has been built, and how to use it, is de
ViViT: Curvature access through the generalized Gauss-Newton's low-rank structure
ViViT is a collection of numerical tricks to efficiently access curvature from the generalized Gauss-Newton (GGN) matrix based on its low-rank structure. Provided functionality includes computing
A small tool to test and visualize protein embeddings and amino acid proportions.
polyprotein_stats A small tool to test and visualize protein embeddings and amino acid proportions. Currently deployed on streamlit.io. Given a set of
Flask Application Structure with MongoDB
This application aims to serve as a template for APIs that intend to use mongoengine and flask-restx

Event queue (Equeue) dialect is an MLIR Dialect that models concurrent devices in terms of control and structure.
Event Queue Dialect Event queue (Equeue) dialect is an MLIR Dialect that models concurrent devices in terms of control and structure. Motivation The m

Source code for EquiDock: Independent SE(3)-Equivariant Models for End-to-End Rigid Protein Docking (ICLR 2022)
Source code for EquiDock: Independent SE(3)-Equivariant Models for End-to-End Rigid Protein Docking (ICLR 2022) Please cite "Independent SE(3)-Equivar
IADS 2021-22 Algorithm and Data structure collection
A collection of algorithms and datastructures introduced during UoE's Introduction to Datastructures and Algorithms class.

OneDriveExplorer - A command line and GUI based application for reconstructing the folder structure of OneDrive from the UserCid.dat file
OneDriveExplorer - A command line and GUI based application for reconstructing the folder structure of OneDrive from the UserCid.dat file

Recovering Brain Structure Network Using Functional Connectivity
Recovering-Brain-Structure-Network-Using-Functional-Connectivity Framework: Papers: This repository provides a PyTorch implementation of the models ad
According to the received excel file (.xlsx,.xlsm,.xltx,.xltm), it converts to word format with a given table structure and formatting
According to the received excel file (.xlsx,.xlsm,.xltx,.xltm), it converts to word format with a given table structure and formatting
UFDR2DIR - A script to convert a Cellebrite UFDR to the original file structure
UFDR2DIR A script to convert a Cellebrite UFDR to it's original file and directo
Drslmarkov - Distributionally Robust Structure Learning for Discrete Pairwise Markov Networks
Distributionally Robust Structure Learning for Discrete Pairwise Markov Networks

SatelliteSfM - A library for solving the satellite structure from motion problem
Satellite Structure from Motion Maintained by Kai Zhang. Overview This is a libr
A data structure that extends pyspark.sql.DataFrame with metadata information.
MetaFrame A data structure that extends pyspark.sql.DataFrame with metadata info
RefineGNN - Iterative refinement graph neural network for antibody sequence-structure co-design (RefineGNN)
Iterative refinement graph neural network for antibody sequence-structure co-des
A scrapy pipeline that provides an easy way to store files and images using various folder structures.
scrapy-folder-tree This is a scrapy pipeline that provides an easy way to store files and images using various folder structures. Supported folder str
Codes and models for the paper "Learning Unknown from Correlations: Graph Neural Network for Inter-novel-protein Interaction Prediction".
GNN_PPI Codes and models for the paper "Learning Unknown from Correlations: Graph Neural Network for Inter-novel-protein Interaction Prediction". Lear

OntoProtein: Protein Pretraining With Ontology Embedding
OntoProtein This is the implement of the paper "OntoProtein: Protein Pretraining With Ontology Embedding". OntoProtein is an effective method that mak
Improved Fitness Optimization Landscapes for Sequence Design
ReLSO Improved Fitness Optimization Landscapes for Sequence Design Description Citation How to run Training models Original data source Description In
CSPML (crystal structure prediction with machine learning-based element substitution)
CSPML (crystal structure prediction with machine learning-based element substitution) CSPML is a unique methodology for the crystal structure predicti
A small automated test structure using python to test *.cpp codes
Get Started Insert C++ Codes Add Test Code Run Test Samples Check Coverages Insert C++ Codes you can easily add c++ files in /inputs directory there i
TGRNet: A Table Graph Reconstruction Network for Table Structure Recognition
TGRNet: A Table Graph Reconstruction Network for Table Structure Recognition Xue, Wenyuan, et al. "TGRNet: A Table Graph Reconstruction Network for Ta
"Structure-Augmented Text Representation Learning for Efficient Knowledge Graph Completion"(WWW 2021)
STAR_KGC This repo contains the source code of the paper accepted by WWW'2021. "Structure-Augmented Text Representation Learning for Efficient Knowled

PyTorch implemention of ICCV'21 paper SGPA: Structure-Guided Prior Adaptation for Category-Level 6D Object Pose Estimation
SGPA: Structure-Guided Prior Adaptation for Category-Level 6D Object Pose Estimation This is the PyTorch implemention of ICCV'21 paper SGPA: Structure
HistoSeg : Quick attention with multi-loss function for multi-structure segmentation in digital histology images
HistoSeg : Quick attention with multi-loss function for multi-structure segmentation in digital histology images Histological Image Segmentation This
PROTEIN EXPRESSION ANALYSIS FOR DOWN SYNDROME
PROTEIN-EXPRESSION-ANALYSIS-FOR-DOWN-SYNDROME Down syndrome (DS) is a chromosomal disorder where organisms have an extra chromosome 21, sometimes know
Tree Nested PyTorch Tensor Lib
DI-treetensor treetensor is a generalized tree-based tensor structure mainly developed by OpenDILab Contributors. Almost all the operation can be supp
Python-boilerplate - Python Boilerplate Project Structure
python-boilerplate Python Boilerplate Project Structure Folder Structure .github
Code repository for Self-supervised Structure-sensitive Learning, CVPR'17
Self-supervised Structure-sensitive Learning (SSL) Ke Gong, Xiaodan Liang, Xiaohui Shen, Liang Lin, "Look into Person: Self-supervised Structure-sensi
RFDesign - Protein hallucination and inpainting with RoseTTAFold
RFDesign: Protein hallucination and inpainting with RoseTTAFold Jue Wang (juewan
💊 A 3D Generative Model for Structure-Based Drug Design (NeurIPS 2021)
A 3D Generative Model for Structure-Based Drug Design Coming soon... Citation @inproceedings{luo2021sbdd, title={A 3D Generative Model for Structu
Toolchain for project structure and documents optimisation
ritocco Toolchain for project structure and documents optimisation
Fairstructure - Structure your data in a FAIR way using google sheets or TSVs
Fairstructure - Structure your data in a FAIR way using google sheets or TSVs. These are then converted to LinkML, and from there other formats
Markov bot - A Writing bot based on Markov Chain for Data Structure Lab
基于马尔可夫链的写作机器人 前端 用html/css完成 Demo展示(已给出文本的相应展示) 用户提供相关的语料库后训练的成果 后端 要完成的几个接口 解析文

BasicNeuralNetwork - This project looks over the basic structure of a neural network and how machine learning training algorithms work
BasicNeuralNetwork - This project looks over the basic structure of a neural network and how machine learning training algorithms work. For this project, I used the sigmoid function as an activation function along with stochastic gradient descent to adjust the weights and biases.

Lolviz - A simple Python data-structure visualization tool for lists of lists, lists, dictionaries; primarily for use in Jupyter notebooks / presentations
lolviz By Terence Parr. See Explained.ai for more stuff. A very nice looking javascript lolviz port with improvements by Adnan M.Sagar. A simple Pytho

Losslandscapetaxonomy - Taxonomizing local versus global structure in neural network loss landscapes
Taxonomizing local versus global structure in neural network loss landscapes Int

PyTorch Lightning + Hydra. A feature-rich template for rapid, scalable and reproducible ML experimentation with best practices. ⚡🔥⚡
Lightning-Hydra-Template A clean and scalable template to kickstart your deep learning project 🚀 ⚡ 🔥 Click on Use this template to initialize new re

Chemical Structure Generator
CSG: Chemical Structure Generator A simple Chemical Structure Generator. Requirements Python 3 (= v3.8) PyQt5 (optional; = v5.15.0 required for grap
A fast Protein Chain / Ligand Extractor and organizer.
Are you tired of using visualization software, or full blown suites just to separate protein chains / ligands ? Are you tired of organizing the mess o

General purpose Slater-Koster tight-binding code for electronic structure calculations
tight-binder Introduction General purpose tight-binding code for electronic structure calculations based on the Slater-Koster approximation. The code

Official Pytorch Implementation of 3DV2021 paper: SAFA: Structure Aware Face Animation.
SAFA: Structure Aware Face Animation (3DV2021) Official Pytorch Implementation of 3DV2021 paper: SAFA: Structure Aware Face Animation. Getting Started
PyTorch code of "SLAPS: Self-Supervision Improves Structure Learning for Graph Neural Networks"
SLAPS-GNN This repo contains the implementation of the model proposed in SLAPS: Self-Supervision Improves Structure Learning for Graph Neural Networks
This program analyzes a DNA sequence and outputs snippets of DNA that are likely to be protein-coding genes.
This program analyzes a DNA sequence and outputs snippets of DNA that are likely to be protein-coding genes.
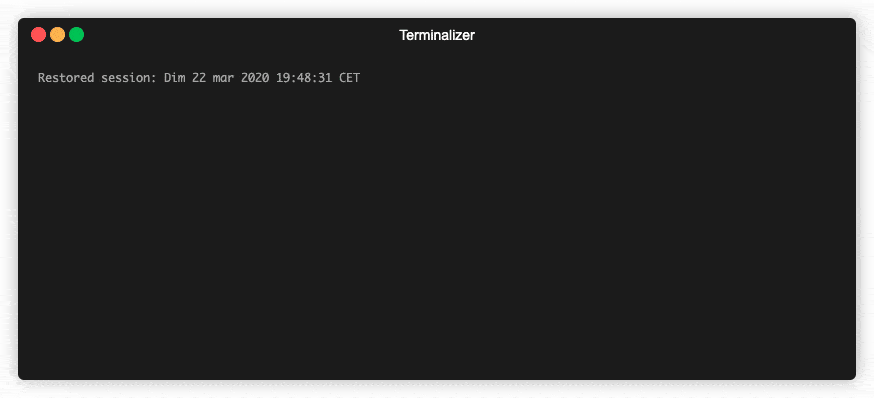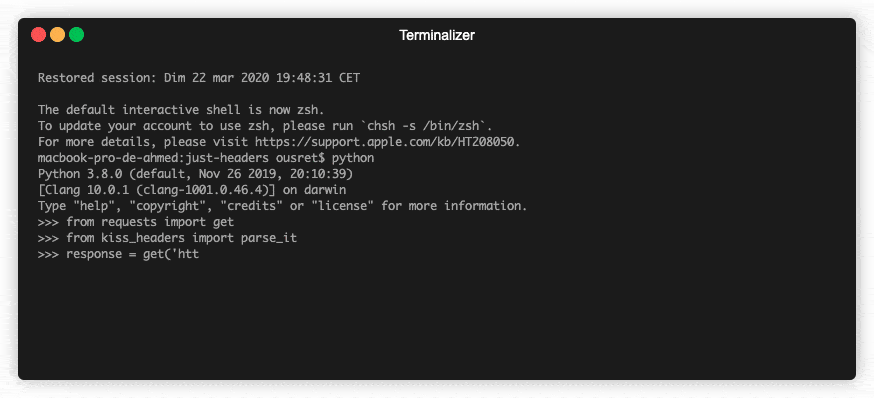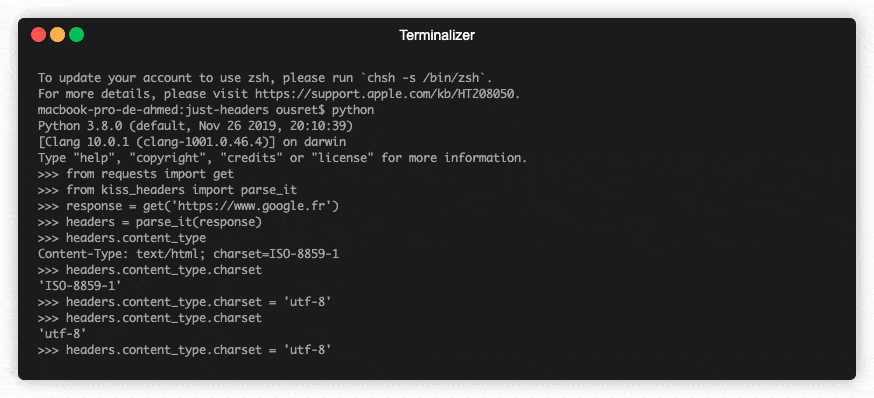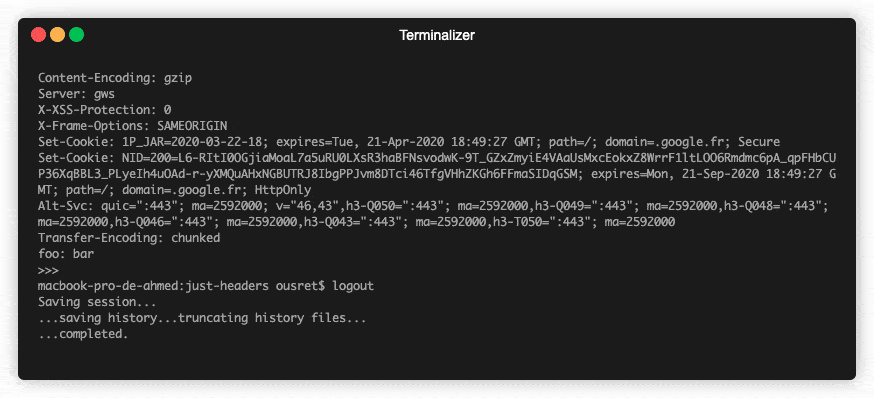
💡Python package for HTTP/1.1 style headers. Parse headers to objects. Most advanced available structure for http headers.
HTTP Headers, the Complete Toolkit 🧰 Object-oriented headers. Kind of structured headers. ❓ Why No matter if you are currently dealing with code usin
Data Structure With Python
Data-Structure-With-Python- Python programs also include in this repo Stack A stack is a linear data structure that stores items in a Last-In/First-Ou

Simple pdf editor while preserving structure and format.
SIMPdf Simple pdf editor while preserving structure and format.
Implementation for paper "STAR: A Structure-aware Lightweight Transformer for Real-time Image Enhancement" (ICCV 2021).
STAR-pytorch Implementation for paper "STAR: A Structure-aware Lightweight Transformer for Real-time Image Enhancement" (ICCV 2021). CVF (pdf) STAR-DC

C3DPO - Canonical 3D Pose Networks for Non-rigid Structure From Motion.
C3DPO: Canonical 3D Pose Networks for Non-Rigid Structure From Motion By: David Novotny, Nikhila Ravi, Benjamin Graham, Natalia Neverova, Andrea Vedal
Official Pytorch implementation of Scene Representation Networks: Continuous 3D-Structure-Aware Neural Scene Representations
Scene Representation Networks This is the official implementation of the NeurIPS submission "Scene Representation Networks: Continuous 3D-Structure-Aw

Infrastructure template and Jupyter notebooks for running RoseTTAFold on AWS Batch.
AWS RoseTTAFold Infrastructure template and Jupyter notebooks for running RoseTTAFold on AWS Batch. Overview Proteins are large biomolecules that play

A Python library for electronic structure pre/post-processing
PyProcar PyProcar is a robust, open-source Python library used for pre- and post-processing of the electronic structure data coming from DFT calculati

An SE(3)-invariant autoencoder for generating the periodic structure of materials
Crystal Diffusion Variational AutoEncoder This software implementes Crystal Diffusion Variational AutoEncoder (CDVAE), which generates the periodic st
![[AAAI 2022] Sparse Structure Learning via Graph Neural Networks for Inductive Document Classification](https://github.com/qkrdmsghk/TextSSL/raw/master/TextSSL.png)
[AAAI 2022] Sparse Structure Learning via Graph Neural Networks for Inductive Document Classification
Sparse Structure Learning via Graph Neural Networks for inductive document classification Make graph dataset create co-occurrence graph for datasets.

The source code of the paper "SHGNN: Structure-Aware Heterogeneous Graph Neural Network"
SHGNN: Structure-Aware Heterogeneous Graph Neural Network The source code and dataset of the paper: SHGNN: Structure-Aware Heterogeneous Graph Neural

CONCEPT (COsmological N-body CodE in PyThon) is a free and open-source simulation code for cosmological structure formation
CONCEPT (COsmological N-body CodE in PyThon) is a free and open-source simulation code for cosmological structure formation. The code should run on any Linux system, from massively parallel computer clusters to laptops.

Implementation of "Semi-supervised Domain Adaptive Structure Learning"
Semi-supervised Domain Adaptive Structure Learning - ASDA This repo contains the source code and dataset for our ASDA paper. Illustration of the propo

A JSON-friendly data structure which allows both object attributes and dictionary keys and values to be used simultaneously and interchangeably.
A JSON-friendly data structure which allows both object attributes and dictionary keys and values to be used simultaneously and interchangeably.
Larch: Applications and Python Library for Data Analysis of X-ray Absorption Spectroscopy (XAS, XANES, XAFS, EXAFS), X-ray Fluorescence (XRF) Spectroscopy and Imaging
Larch: Data Analysis Tools for X-ray Spectroscopy and More Documentation: http://xraypy.github.io/xraylarch Code: http://github.com/xraypy/xraylarch L

Simple Tensorflow implementation of "Adaptive Convolutions for Structure-Aware Style Transfer" (CVPR 2021)
AdaConv — Simple TensorFlow Implementation [Paper] : Adaptive Convolutions for Structure-Aware Style Transfer (CVPR 2021) Note This repository does no
Uni-Fold: Training your own deep protein-folding models.
Uni-Fold: Training your own deep protein-folding models. This package provides and implementation of a trainable, Transformer-based deep protein foldi
Garbage classification using structure data.
垃圾分类模型使用说明 1.包含以下数据文件 文件 描述 data/MaterialMapping.csv 物体以及其归类的信息 data/TestRecords 光谱原始测试数据 CSV 文件 data/TestRecordDesc.zip CSV 文件描述文件 data/Boundaries.cs
Uni-Fold: Training your own deep protein-folding models
Uni-Fold: Training your own deep protein-folding models. This package provides an implementation of a trainable, Transformer-based deep protein foldin

Implementation of the ivis algorithm as described in the paper Structure-preserving visualisation of high dimensional single-cell datasets.
Implementation of the ivis algorithm as described in the paper Structure-preserving visualisation of high dimensional single-cell datasets.
Flask pre-setup architecture. This can be used in any flask project for a faster and better project code structure.
Flask pre-setup architecture. This can be used in any flask project for a faster and better project code structure. All the required libraries are already installed easily to use in any big project.
Codebase for Inducing Causal Structure for Interpretable Neural Networks
Interchange Intervention Training (IIT) Codebase for Inducing Causal Structure for Interpretable Neural Networks Release Notes 12/01/2021: Code and Pa

My notes on Data structure and Algos in golang implementation and python
My notes on DS and Algo Table of Contents Arrays LinkedList Trees Types of trees: Tree/Graph Traversal Algorithms Heap Priorty Queue Trie Graphs Graph

Implementation of "Semi-supervised Domain Adaptive Structure Learning"
Semi-supervised Domain Adaptive Structure Learning - ASDA This repo contains the source code and dataset for our ASDA paper. Illustration of the propo

The source code of "SIDE: Center-based Stereo 3D Detector with Structure-aware Instance Depth Estimation", accepted to WACV 2022.
SIDE: Center-based Stereo 3D Detector with Structure-aware Instance Depth Estimation The source code of our work "SIDE: Center-based Stereo 3D Detecto

Making Structure-from-Motion (COLMAP) more robust to symmetries and duplicated structures
SfM disambiguation with COLMAP About Structure-from-Motion generally fails when the scene exhibits symmetries and duplicated structures. In this repos

Decoding the Protein-ligand Interactions Using Parallel Graph Neural Networks
Decoding the Protein-ligand Interactions Using Parallel Graph Neural Networks Requirements python 0.10+ rdkit 2020.03.3.0 biopython 1.78 openbabel 2.4

This tool is designed to help administrators get an overview of their Active Directory structure.
This tool is designed to help administrators get an overview of their Active Directory structure. In the group view you can see all elements of an AD (OU, USER, GROUPS, COMPUTERS etc.). In the user view one sees all users according to their Manager/Manager of Hierarchy.
Repository containing documentation about the Minecraft Legacy FUI file structure
Repository containing documentation about the Minecraft Legacy FUI file structure
A logical, reasonably standardized, but flexible project structure for doing and sharing data science work.
Cookiecutter Data Science A logical, reasonably standardized, but flexible project structure for doing and sharing data science work. Project homepage

A Protein-RNA Interface Predictor Based on Semantics of Sequences
PRIP PRIP:A Protein-RNA Interface Predictor Based on Semantics of Sequences installation gensim==3.8.3 matplotlib==3.1.3 xgboost==1.3.3 prettytable==2

An implementation for the ICCV 2021 paper Deep Permutation Equivariant Structure from Motion.
Deep Permutation Equivariant Structure from Motion Paper | Poster This repository contains an implementation for the ICCV 2021 paper Deep Permutation
A package to predict protein inter-residue geometries from sequence data
trRosetta This package is a part of trRosetta protein structure prediction protocol developed in: Improved protein structure prediction using predicte
COLMAP - Structure-from-Motion and Multi-View Stereo
COLMAP About COLMAP is a general-purpose Structure-from-Motion (SfM) and Multi-View Stereo (MVS) pipeline with a graphical and command-line interface.

Geometric Vector Perceptrons --- a rotation-equivariant GNN for learning from biomolecular structure
Geometric Vector Perceptron Implementation of equivariant GVP-GNNs as described in Learning from Protein Structure with Geometric Vector Perceptrons b
A Python Tools to imaging the shallow seismic structure
ShallowSeismicImaging Tools to imaging the shallow seismic structure, above 10 km, based on the ZH ratio measured from the ambient seismic noise, and

Bio-Computing Platform Featuring Large-Scale Representation Learning and Multi-Task Deep Learning “螺旋桨”生物计算工具集
English | 简体中文 Latest News 2021.10.25 Paper "Docking-based Virtual Screening with Multi-Task Learning" is accepted by BIBM 2021. 2021.07.29 PaddleHeli
AptaMat is a simple script which aims to measure differences between DNA or RNA secondary structures.
AptaMAT Purpose AptaMat is a simple script which aims to measure differences between DNA or RNA secondary structures. The method is based on the compa
Code for generating alloy / disordered structures through the special quasirandom structure (SQS) algorithm
Code for generating alloy / disordered structures through the special quasirandom structure (SQS) algorithm

Erosion and dialation using structure element in OpenCV python
Erosion and dialation using structure element in OpenCV python

Python program for analyzing the output files of phonopy.
PhononTools Description Python program to analyze the results generated by phonopy. Using the .yaml and .dat files that phonopy generates one can plot

This package is a python library with tools for the Molecular Simulation - Software Gromos.
This package is a python library with tools for the Molecular Simulation - Software Gromos. It allows you to easily set up, manage and analyze simulations in python.