112 Repositories
Python protein-protein-interaction Libraries

Jupyter Dock is a set of Jupyter Notebooks for performing molecular docking protocols interactively, as well as visualizing, converting file formats and analyzing the results.
Molecular Docking integrated in Jupyter Notebooks Description | Citation | Installation | Examples | Limitations | License Table of content Descriptio

RITA is a family of autoregressive protein models, developed by LightOn in collaboration with the OATML group at Oxford and the Debora Marks Lab at Harvard.
RITA: a Study on Scaling Up Generative Protein Sequence Models RITA is a family of autoregressive protein models, developed by a collaboration of Ligh

TigerLily: Finding drug interactions in silico with the Graph.
Drug Interaction Prediction with Tigerlily Documentation | Example Notebook | Youtube Video | Project Report Tigerlily is a TigerGraph based system de

Code for Ditto: Building Digital Twins of Articulated Objects from Interaction
Ditto: Building Digital Twins of Articulated Objects from Interaction Zhenyu Jiang, Cheng-Chun Hsu, Yuke Zhu CVPR 2022, Oral Project | arxiv News 2022

Code for our CVPR 2022 Paper "GEN-VLKT: Simplify Association and Enhance Interaction Understanding for HOI Detection"
GEN-VLKT Code for our CVPR 2022 paper "GEN-VLKT: Simplify Association and Enhance Interaction Understanding for HOI Detection". Contributed by Yue Lia

Implementation of a protein autoregressive language model, but with autoregressive infilling objective (editing subsequences capability)
Protein GLM (wip) Implementation of a protein autoregressive language model, but with autoregressive infilling objective (editing subsequences capabil

Implementation of the GVP-Transformer, which was used in the paper "Learning inverse folding from millions of predicted structures" for de novo protein design alongside Alphafold2
GVP Transformer (wip) Implementation of the GVP-Transformer, which was used in the paper Learning inverse folding from millions of predicted structure

Official implementation of "Generating 3D Molecules for Target Protein Binding"
Generating 3D Molecules for Target Protein Binding This is the official implementation of the GraphBP method proposed in the following paper. Meng Liu

Addon and nodes for working with structural biology and molecular data in Blender.
Molecular Nodes 🧬 🔬 💻 Buy Me a Coffee to Keep Development Going! Join a Community of Blender SciVis People! What is Molecular Nodes? Molecular Node

ProtFeat is protein feature extraction tool that utilizes POSSUM and iFeature.
Description: ProtFeat is designed to extract the protein features by employing POSSUM and iFeature python-based tools. ProtFeat includes a total of 39
![[arXiv22] Disentangled Representation Learning for Text-Video Retrieval](https://github.com/foolwood/DRL/raw/main/demo/pipeline.png)
[arXiv22] Disentangled Representation Learning for Text-Video Retrieval
Disentangled Representation Learning for Text-Video Retrieval This is a PyTorch implementation of the paper Disentangled Representation Learning for T

EquiBind: Geometric Deep Learning for Drug Binding Structure Prediction
EquiBind: geometric deep learning for fast predictions of the 3D structure in which a small molecule binds to a protein

Benchmarking Pipeline for Prediction of Protein-Protein Interactions
B4PPI Benchmarking Pipeline for the Prediction of Protein-Protein Interactions How this benchmarking pipeline has been built, and how to use it, is de
A small tool to test and visualize protein embeddings and amino acid proportions.
polyprotein_stats A small tool to test and visualize protein embeddings and amino acid proportions. Currently deployed on streamlit.io. Given a set of

Detecting Human-Object Interactions with Object-Guided Cross-Modal Calibrated Semantics
[AAAI2022] Detecting Human-Object Interactions with Object-Guided Cross-Modal Calibrated Semantics Overall pipeline of OCN. Paper Link: [arXiv] [AAAI

Source code for EquiDock: Independent SE(3)-Equivariant Models for End-to-End Rigid Protein Docking (ICLR 2022)
Source code for EquiDock: Independent SE(3)-Equivariant Models for End-to-End Rigid Protein Docking (ICLR 2022) Please cite "Independent SE(3)-Equivar
Spectral decomposition for characterizing long-range interaction profiles in Hi-C maps
Inspectral Spectral decomposition for characterizing long-range interaction prof
RDMAss - A Python Discord bot creating an interaction with RDM API
RDMAss A Python Discord bot creating an interaction with RDM API. Features Assig
Codes and models for the paper "Learning Unknown from Correlations: Graph Neural Network for Inter-novel-protein Interaction Prediction".
GNN_PPI Codes and models for the paper "Learning Unknown from Correlations: Graph Neural Network for Inter-novel-protein Interaction Prediction". Lear

OntoProtein: Protein Pretraining With Ontology Embedding
OntoProtein This is the implement of the paper "OntoProtein: Protein Pretraining With Ontology Embedding". OntoProtein is an effective method that mak
Improved Fitness Optimization Landscapes for Sequence Design
ReLSO Improved Fitness Optimization Landscapes for Sequence Design Description Citation How to run Training models Original data source Description In

PyTorch implementation of the ideas presented in the paper Interaction Grounded Learning (IGL)
Interaction Grounded Learning This repository contains a simple PyTorch implementation of the ideas presented in the paper Interaction Grounded Learni
PROTEIN EXPRESSION ANALYSIS FOR DOWN SYNDROME
PROTEIN-EXPRESSION-ANALYSIS-FOR-DOWN-SYNDROME Down syndrome (DS) is a chromosomal disorder where organisms have an extra chromosome 21, sometimes know

Full Transformer Framework for Robust Point Cloud Registration with Deep Information Interaction
Full Transformer Framework for Robust Point Cloud Registration with Deep Information Interaction. arxiv This repository contains python scripts for tr

As a part of the HAKE project, includes the reproduced SOTA models and the corresponding HAKE-enhanced versions (CVPR2020).
HAKE-Action HAKE-Action (TensorFlow) is a project to open the SOTA action understanding studies based on our Human Activity Knowledge Engine. It inclu
RFDesign - Protein hallucination and inpainting with RoseTTAFold
RFDesign: Protein hallucination and inpainting with RoseTTAFold Jue Wang (juewan

This repository is for our paper Exploiting Scene Graphs for Human-Object Interaction Detection accepted by ICCV 2021.
SG2HOI This repository is for our paper Exploiting Scene Graphs for Human-Object Interaction Detection accepted by ICCV 2021. Installation Pytorch 1.7

A web-based analysis toolkit for the System Usability Scale providing calculation, plotting, interpretation and contextualization utility
System Usability Scale Analysis Toolkit The System Usability Scale (SUS) Analysis Toolkit is a web-based python application that provides a compilatio

NiceGUI is an easy to use, Python-based UI framework, which renderes to the web browser.
NiceGUI NiceGUI is an easy to use, Python-based UI framework, which renderes to the web browser. You can create buttons, dialogs, markdown, 3D scences

A discord.py extension for sending, receiving and handling ui interactions in discord
discord-ui A discord.py extension for using discord ui/interaction features pip package ▪ read the docs ▪ examples Introduction This is a discord.py u
A fast Protein Chain / Ligand Extractor and organizer.
Are you tired of using visualization software, or full blown suites just to separate protein chains / ligands ? Are you tired of organizing the mess o
This program analyzes a DNA sequence and outputs snippets of DNA that are likely to be protein-coding genes.
This program analyzes a DNA sequence and outputs snippets of DNA that are likely to be protein-coding genes.

A PyTorch based deep learning library for drug pair scoring.
Documentation | External Resources | Datasets | Examples ChemicalX is a deep learning library for drug-drug interaction, polypharmacy side effect and
An experimentation and research platform to investigate the interaction of automated agents in an abstract simulated network environments.
CyberBattleSim April 8th, 2021: See the announcement on the Microsoft Security Blog. CyberBattleSim is an experimentation research platform to investi

Real-Time and Accurate Full-Body Multi-Person Pose Estimation&Tracking System
News! Aug 2020: v0.4.0 version of AlphaPose is released! Stronger tracking! Include whole body(face,hand,foot) keypoints! Colab now available. Dec 201
Codes for building and training the neural network model described in Domain-informed neural networks for interaction localization within astroparticle experiments.
Domain-informed Neural Networks Codes for building and training the neural network model described in Domain-informed neural networks for interaction

QAHOI: Query-Based Anchors for Human-Object Interaction Detection (paper)
QAHOI QAHOI: Query-Based Anchors for Human-Object Interaction Detection (paper) Requirements PyTorch = 1.5.1 torchvision = 0.6.1 pip install -r requ

Infrastructure template and Jupyter notebooks for running RoseTTAFold on AWS Batch.
AWS RoseTTAFold Infrastructure template and Jupyter notebooks for running RoseTTAFold on AWS Batch. Overview Proteins are large biomolecules that play
People Interaction Graph
Gihan Jayatilaka*, Jameel Hassan*, Suren Sritharan*, Janith Senananayaka, Harshana Weligampola, et. al., 2021. Holistic Interpretation of Public Scenes Using Computer Vision and Temporal Graphs to Identify Social Distancing Violations. arXiv preprint.
POC for detecting the Log4Shell (Log4J RCE) vulnerability
Interactsh An OOB interaction gathering server and client library Features • Usage • Interactsh Client • Interactsh Server • Interactsh Integration •

Anchor Retouching via Model Interaction for Robust Object Detection in Aerial Images
Anchor Retouching via Model Interaction for Robust Object Detection in Aerial Images In this paper, we present an effective Dynamic Enhancement Anchor
Uni-Fold: Training your own deep protein-folding models.
Uni-Fold: Training your own deep protein-folding models. This package provides and implementation of a trainable, Transformer-based deep protein foldi
Uni-Fold: Training your own deep protein-folding models
Uni-Fold: Training your own deep protein-folding models. This package provides an implementation of a trainable, Transformer-based deep protein foldin

ColBERT: Contextualized Late Interaction over BERT (SIGIR'20)
Update: if you're looking for ColBERTv2 code, you can find it alongside a new simpler API, in the branch new_api. ColBERT ColBERT is a fast and accura

Official PyTorch implementation for paper "Efficient Two-Stage Detection of Human–Object Interactions with a Novel Unary–Pairwise Transformer"
UPT: Unary–Pairwise Transformers This repository contains the official PyTorch implementation for the paper Frederic Z. Zhang, Dylan Campbell and Step

Official Pytorch implementation for 2021 ICCV paper "Learning Motion Priors for 4D Human Body Capture in 3D Scenes" and trained models / data
Learning Motion Priors for 4D Human Body Capture in 3D Scenes (LEMO) Official Pytorch implementation for 2021 ICCV (oral) paper "Learning Motion Prior

This repository provides code for "On Interaction Between Augmentations and Corruptions in Natural Corruption Robustness".
On Interaction Between Augmentations and Corruptions in Natural Corruption Robustness This repository provides the code for the paper On Interaction B
Graph Self-Attention Network for Learning Spatial-Temporal Interaction Representation in Autonomous Driving
GSAN Introduction Code for paper GSAN: Graph Self-Attention Network for Learning Spatial-Temporal Interaction Representation in Autonomous Driving, wh

Decoding the Protein-ligand Interactions Using Parallel Graph Neural Networks
Decoding the Protein-ligand Interactions Using Parallel Graph Neural Networks Requirements python 0.10+ rdkit 2020.03.3.0 biopython 1.78 openbabel 2.4
Multi-modal co-attention for drug-target interaction annotation and Its Application to SARS-CoV-2
CoaDTI Multi-modal co-attention for drug-target interaction annotation and Its Application to SARS-CoV-2 Abstract Environment The test was conducted i

A Protein-RNA Interface Predictor Based on Semantics of Sequences
PRIP PRIP:A Protein-RNA Interface Predictor Based on Semantics of Sequences installation gensim==3.8.3 matplotlib==3.1.3 xgboost==1.3.3 prettytable==2
A package to predict protein inter-residue geometries from sequence data
trRosetta This package is a part of trRosetta protein structure prediction protocol developed in: Improved protein structure prediction using predicte

Bio-Computing Platform Featuring Large-Scale Representation Learning and Multi-Task Deep Learning “螺旋桨”生物计算工具集
English | 简体中文 Latest News 2021.10.25 Paper "Docking-based Virtual Screening with Multi-Task Learning" is accepted by BIBM 2021. 2021.07.29 PaddleHeli
Ingeniamotion is a library that works over ingenialink and aims to simplify the interaction with Ingenia's drives.
Ingeniamotion Ingeniamotion is a library that works over ingenialink and aims to simplify the interaction with Ingenia's drives. Requirements Python 3
AIR^2 for Interaction Prediction
This is the repository for AIR^2 for Interaction Prediction. Explanation of the solution: Video: link License AIR is released under the Apache 2.0 lic
Visual Interaction with Code - A portable visual debugger for python
VIC Visual Interaction with Code A simple tool for debugging and interacting with running python code. This tool is designed to make it easy to inspec

Readings for "A Unified View of Relational Deep Learning for Polypharmacy Side Effect, Combination Therapy, and Drug-Drug Interaction Prediction."
Polypharmacy - DDI - Synergy Survey The Survey Paper This repository accompanies our survey paper A Unified View of Relational Deep Learning for Polyp

This package is a python library with tools for the Molecular Simulation - Software Gromos.
This package is a python library with tools for the Molecular Simulation - Software Gromos. It allows you to easily set up, manage and analyze simulations in python.
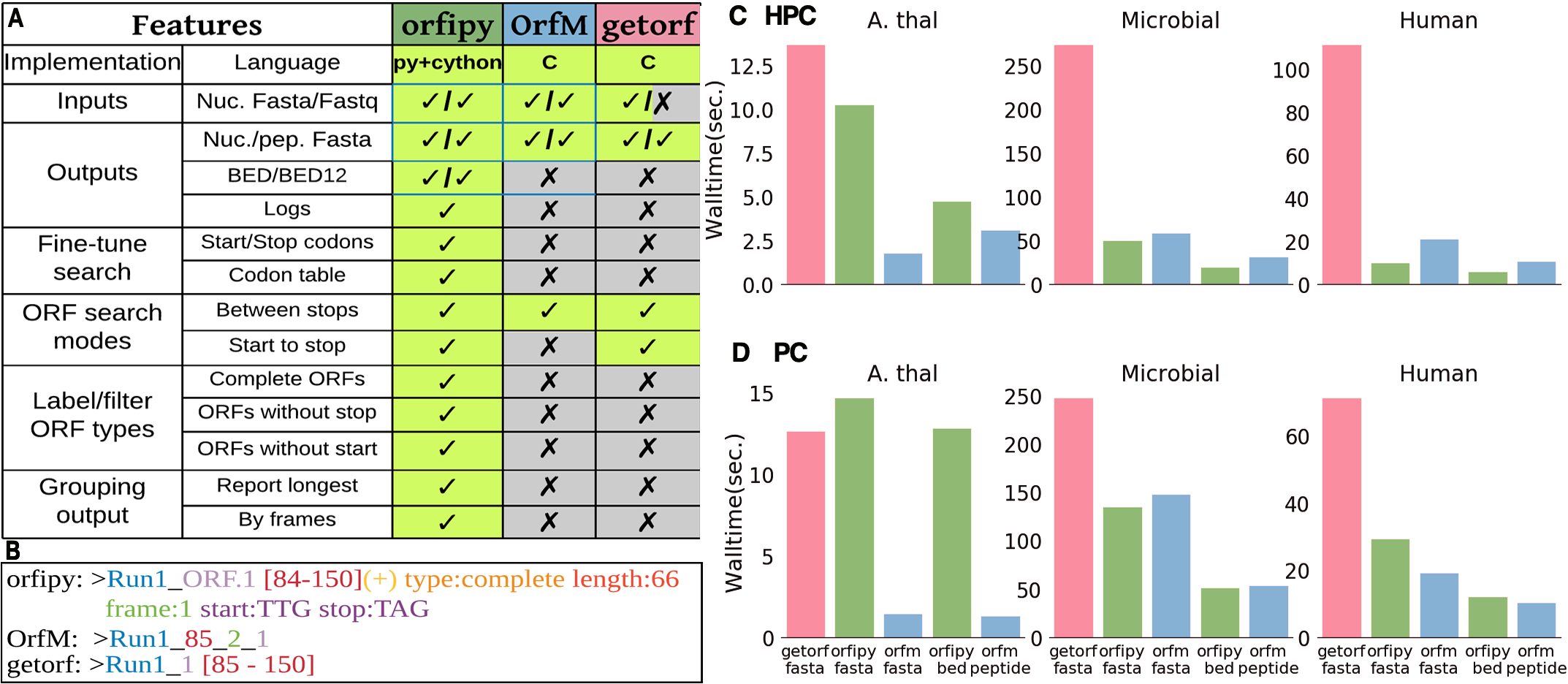
orfipy is a tool written in python/cython to extract ORFs in an extremely and fast and flexible manner
Introduction orfipy is a tool written in python/cython to extract ORFs in an extremely and fast and flexible manner. Other popular ORF searching tools

cLoops2: full stack analysis tool for chromatin interactions
cLoops2: full stack analysis tool for chromatin interactions Introduction cLoops2 is an extension of our previous work, cLoops. From loop-calling base

Bioinformatics tool for exploring RNA-Protein interactions
Explore RNA-Protein interactions. RNPFind is a bioinformatics tool. It takes an RNA transcript as input and gives a list of RNA binding protein (RBP)

Readings for "A Unified View of Relational Deep Learning for Polypharmacy Side Effect, Combination Therapy, and Drug-Drug Interaction Prediction."
Polypharmacy - DDI - Synergy Survey The Survey Paper This repository accompanies our survey paper A Unified View of Relational Deep Learning for Polyp
Allows simplified Python interaction with Rapid7's InsightIDR REST API.
InsightIDR4Py Allows simplified Python interaction with Rapid7's InsightIDR REST API. InsightIDR4Py allows analysts to query log data from Rapid7 Insi
Deep generative models of 3D grids for structure-based drug discovery
What is liGAN? liGAN is a research codebase for training and evaluating deep generative models for de novo drug design based on 3D atomic density grid
Predicting lncRNA–protein interactions based on graph autoencoders and collaborative training
Predicting lncRNA–protein interactions based on graph autoencoders and collaborative training Code for our paper "Predicting lncRNA–protein interactio
peptides.py is a pure-Python package to compute common descriptors for protein sequences
peptides.py Physicochemical properties and indices for amino-acid sequences. 🗺️ Overview peptides.py is a pure-Python package to compute common descr
AEI: Actors-Environment Interaction with Adaptive Attention for Temporal Action Proposals Generation
AEI: Actors-Environment Interaction with Adaptive Attention for Temporal Action Proposals Generation A pytorch-version implementation codes of paper:
GPT-3 command line interaction
Writer_unblock Straight-forward command line interfacing with GPT-3. Finding yourself stuck at a conceptual stage? Spinning your wheels needlessly on

Official implementation for Multi-Modal Interaction Graph Convolutional Network for Temporal Language Localization in Videos
Multi-modal Interaction Graph Convolutioal Network for Temporal Language Localization in Videos Official implementation for Multi-Modal Interaction Gr
A graph neural network (GNN) model to predict protein-protein interactions (PPI) with no sample features
A graph neural network (GNN) model to predict protein-protein interactions (PPI) with no sample features
A geometric deep learning pipeline for predicting protein interface contacts.
A geometric deep learning pipeline for predicting protein interface contacts.
The GitHub repository for the paper: “Time Series is a Special Sequence: Forecasting with Sample Convolution and Interaction“.
SCINet This is the original PyTorch implementation of the following work: Time Series is a Special Sequence: Forecasting with Sample Convolution and I
Replication attempt for the Protein Folding Model
RGN2-Replica (WIP) To eventually become an unofficial working Pytorch implementation of RGN2, an state of the art model for MSA-less Protein Folding f

GBIM(Gesture-Based Interaction map)
手势交互地图 GBIM(Gesture-Based Interaction map),基于视觉深度神经网络的交互地图,通过电脑摄像头观察使用者的手势变化,进而控制地图进行简单的交互。网络使用PaddleX提供的轻量级模型PPYOLO Tiny以及MobileNet V3 small,使得整个模型大小约10MB左右,即使在CPU下也能快速定位和识别手势。

Graph-based community clustering approach to extract protein domains from a predicted aligned error matrix
Using a predicted aligned error matrix corresponding to an AlphaFold2 model , returns a series of lists of residue indices, where each list corresponds to a set of residues clustering together into a pseudo-rigid domain.
A python discord client interaction emulator for the DC29 badge code channel
dc29-discord-signalbot A python discord client interaction emulator for the DC29 badge code channel Prep Open Developer mode Open the developer mode f
Pytorch Implementation of Interaction Networks for Learning about Objects, Relations and Physics
Interaction-Network-Pytorch Pytorch Implementraion of Interaction Networks for Learning about Objects, Relations and Physics. Interaction Network is a

This's an implementation of deepmind Visual Interaction Networks paper using pytorch
Visual-Interaction-Networks An implementation of Deepmind visual interaction networks in Pytorch. Introduction For the purpose of understanding the ch

Implementation of the Triangle Multiplicative module, used in Alphafold2 as an efficient way to mix rows or columns of a 2d feature map, as a standalone package for Pytorch
Triangle Multiplicative Module - Pytorch Implementation of the Triangle Multiplicative module, used in Alphafold2 as an efficient way to mix rows or c

Implementation of Invariant Point Attention, used for coordinate refinement in the structure module of Alphafold2, as a standalone Pytorch module
Invariant Point Attention - Pytorch Implementation of Invariant Point Attention as a standalone module, which was used in the structure module of Alph
WILSON Cloud Respwnder is a Web Interaction Logger Sending Out Notifications with the ability to serve custom content in order to appropriately respond to client-issued requests.
WILSON Cloud Respwnder What is this? WILSON Cloud Respwnder is a Web Interaction Logger Sending Out Notifications (WILSON) with the ability to serve c

Code for KDD'20 "An Efficient Neighborhood-based Interaction Model for Recommendation on Heterogeneous Graph"
Heterogeneous INteract and aggreGatE (GraphHINGE) This is a pytorch implementation of GraphHINGE model. This is the experiment code in the following w
Implementation and replication of ProGen, Language Modeling for Protein Generation, in Jax
ProGen - (wip) Implementation and replication of ProGen, Language Modeling for Protein Generation, in Pytorch and Jax (the weights will be made easily
7th place solution of Human Protein Atlas - Single Cell Classification on Kaggle
kaggle-hpa-2021-7th-place-solution Code for 7th place solution of Human Protein Atlas - Single Cell Classification on Kaggle. A description of the met

A denoising diffusion probabilistic model (DDPM) tailored for conditional generation of protein distograms
Denoising Diffusion Probabilistic Model for Proteins Implementation of Denoising Diffusion Probabilistic Model in Pytorch. It is a new approach to gen

Unofficial TensorFlow implementation of Protein Interface Prediction using Graph Convolutional Networks.
[TensorFlow] Protein Interface Prediction using Graph Convolutional Networks Unofficial TensorFlow implementation of Protein Interface Prediction usin

Official repository for HOTR: End-to-End Human-Object Interaction Detection with Transformers (CVPR'21, Oral Presentation)
Official PyTorch Implementation for HOTR: End-to-End Human-Object Interaction Detection with Transformers (CVPR'2021, Oral Presentation) HOTR: End-to-

This is the research repository for Vid2Doppler: Synthesizing Doppler Radar Data from Videos for Training Privacy-Preserving Activity Recognition.
Vid2Doppler: Synthesizing Doppler Radar Data from Videos for Training Privacy-Preserving Activity Recognition This is the research repository for Vid2
Protein Language Model
ProteinLM We pretrain protein language model based on Megatron-LM framework, and then evaluate the pretrained model results on TAPE (Tasks Assessing P
PIGLeT: Language Grounding Through Neuro-Symbolic Interaction in a 3D World [ACL 2021]
piglet PIGLeT: Language Grounding Through Neuro-Symbolic Interaction in a 3D World [ACL 2021] This repo contains code and data for PIGLeT. If you like
Implementation of ProteinBERT in Pytorch
ProteinBERT - Pytorch (wip) Implementation of ProteinBERT in Pytorch. Original Repository Install $ pip install protein-bert-pytorch Usage import torc

NExT-QA: Next Phase of Question-Answering to Explaining Temporal Actions (CVPR2021)
NExT-QA We reproduce some SOTA VideoQA methods to provide benchmark results for our NExT-QA dataset accepted to CVPR2021 (with 1 'Strong Accept' and 2
This is the repo for the paper `SumGNN: Multi-typed Drug Interaction Prediction via Efficient Knowledge Graph Summarization'. (published in Bioinformatics'21)
SumGNN: Multi-typed Drug Interaction Prediction via Efficient Knowledge Graph Summarization This is the code for our paper ``SumGNN: Multi-typed Drug
Implementation of ProteinBERT in Pytorch
ProteinBERT - Pytorch (wip) Implementation of ProteinBERT in Pytorch. Original Repository Install $ pip install protein-bert-pytorch Usage import torc
ProteinBERT is a universal protein language model pretrained on ~106M proteins from the UniRef90 dataset.
ProteinBERT is a universal protein language model pretrained on ~106M proteins from the UniRef90 dataset. Through its Python API, the pretrained model can be fine-tuned on any protein-related task in a matter of minutes. Based on our experiments with a wide range of benchmarks, ProteinBERT usually achieves state-of-the-art performance. ProteinBERT is built on TenforFlow/Keras.

Deep functional residue identification
DeepFRI Deep functional residue identification Citing @article {Gligorijevic2019, author = {Gligorijevic, Vladimir and Renfrew, P. Douglas and Koscio

Populating 3D Scenes by Learning Human-Scene Interaction https://posa.is.tue.mpg.de/
Populating 3D Scenes by Learning Human-Scene Interaction [Project Page] [Paper] License Software Copyright License for non-commercial scientific resea

Generative Models for Graph-Based Protein Design
Graph-Based Protein Design This repo contains code for Generative Models for Graph-Based Protein Design by John Ingraham, Vikas Garg, Regina Barzilay
Structural basis for solubility in protein expression systems
Structural basis for solubility in protein expression systems Large-scale protein production for biotechnology and biopharmaceutical applications rely
