100 Repositories
Python protein-protein-interactions Libraries

Jupyter Dock is a set of Jupyter Notebooks for performing molecular docking protocols interactively, as well as visualizing, converting file formats and analyzing the results.
Molecular Docking integrated in Jupyter Notebooks Description | Citation | Installation | Examples | Limitations | License Table of content Descriptio

RITA is a family of autoregressive protein models, developed by LightOn in collaboration with the OATML group at Oxford and the Debora Marks Lab at Harvard.
RITA: a Study on Scaling Up Generative Protein Sequence Models RITA is a family of autoregressive protein models, developed by a collaboration of Ligh

TigerLily: Finding drug interactions in silico with the Graph.
Drug Interaction Prediction with Tigerlily Documentation | Example Notebook | Youtube Video | Project Report Tigerlily is a TigerGraph based system de

Code Implementation of "Learning Span-Level Interactions for Aspect Sentiment Triplet Extraction".
Span-ASTE: Learning Span-Level Interactions for Aspect Sentiment Triplet Extraction ***** New March 31th, 2022: Scikit-Style API for Easy Usage *****
CompleX Group Interactions (XGI) provides an ecosystem for the analysis and representation of complex systems with group interactions.
XGI CompleX Group Interactions (XGI) is a Python package for the representation, manipulation, and study of the structure, dynamics, and functions of

Implementation of a protein autoregressive language model, but with autoregressive infilling objective (editing subsequences capability)
Protein GLM (wip) Implementation of a protein autoregressive language model, but with autoregressive infilling objective (editing subsequences capabil

Implementation of the GVP-Transformer, which was used in the paper "Learning inverse folding from millions of predicted structures" for de novo protein design alongside Alphafold2
GVP Transformer (wip) Implementation of the GVP-Transformer, which was used in the paper Learning inverse folding from millions of predicted structure

Official implementation of "Generating 3D Molecules for Target Protein Binding"
Generating 3D Molecules for Target Protein Binding This is the official implementation of the GraphBP method proposed in the following paper. Meng Liu

Addon and nodes for working with structural biology and molecular data in Blender.
Molecular Nodes 🧬 🔬 💻 Buy Me a Coffee to Keep Development Going! Join a Community of Blender SciVis People! What is Molecular Nodes? Molecular Node

ProtFeat is protein feature extraction tool that utilizes POSSUM and iFeature.
Description: ProtFeat is designed to extract the protein features by employing POSSUM and iFeature python-based tools. ProtFeat includes a total of 39

Repository for "Improving evidential deep learning via multi-task learning," published in AAAI2022
Improving evidential deep learning via multi task learning It is a repository of AAAI2022 paper, “Improving evidential deep learning via multi-task le

EquiBind: Geometric Deep Learning for Drug Binding Structure Prediction
EquiBind: geometric deep learning for fast predictions of the 3D structure in which a small molecule binds to a protein
Negative Interactions for Improved Collaborative Filtering:
Negative Interactions for Improved Collaborative Filtering: Don’t go Deeper, go Higher This notebook provides an implementation in Python 3 of the alg

Benchmarking Pipeline for Prediction of Protein-Protein Interactions
B4PPI Benchmarking Pipeline for the Prediction of Protein-Protein Interactions How this benchmarking pipeline has been built, and how to use it, is de
A small tool to test and visualize protein embeddings and amino acid proportions.
polyprotein_stats A small tool to test and visualize protein embeddings and amino acid proportions. Currently deployed on streamlit.io. Given a set of

Detecting Human-Object Interactions with Object-Guided Cross-Modal Calibrated Semantics
[AAAI2022] Detecting Human-Object Interactions with Object-Guided Cross-Modal Calibrated Semantics Overall pipeline of OCN. Paper Link: [arXiv] [AAAI

Source code for EquiDock: Independent SE(3)-Equivariant Models for End-to-End Rigid Protein Docking (ICLR 2022)
Source code for EquiDock: Independent SE(3)-Equivariant Models for End-to-End Rigid Protein Docking (ICLR 2022) Please cite "Independent SE(3)-Equivar
Codes and models for the paper "Learning Unknown from Correlations: Graph Neural Network for Inter-novel-protein Interaction Prediction".
GNN_PPI Codes and models for the paper "Learning Unknown from Correlations: Graph Neural Network for Inter-novel-protein Interaction Prediction". Lear

OntoProtein: Protein Pretraining With Ontology Embedding
OntoProtein This is the implement of the paper "OntoProtein: Protein Pretraining With Ontology Embedding". OntoProtein is an effective method that mak
Improved Fitness Optimization Landscapes for Sequence Design
ReLSO Improved Fitness Optimization Landscapes for Sequence Design Description Citation How to run Training models Original data source Description In
A secure and customizable bot for controlling cross-server announcements and interactions within Discord
DiscordBot A secure and customizable bot for controlling cross-server announcements and interactions within Discord. Within the code of the bot, you c
PROTEIN EXPRESSION ANALYSIS FOR DOWN SYNDROME
PROTEIN-EXPRESSION-ANALYSIS-FOR-DOWN-SYNDROME Down syndrome (DS) is a chromosomal disorder where organisms have an extra chromosome 21, sometimes know
A sample pytorch Implementation of ACL 2021 research paper "Learning Span-Level Interactions for Aspect Sentiment Triplet Extraction".
Span-ASTE-Pytorch This repository is a pytorch version that implements Ali's ACL 2021 research paper Learning Span-Level Interactions for Aspect Senti
Message commands extension for discord-py-interactions
interactions-message-commands Message commands extension for discord-py-interactions README IS NOT FINISHED YET BUT IT IS A GOOD START Installation pi

2021:"Bridging Global Context Interactions for High-Fidelity Image Completion"
TFill arXiv | Project This repository implements the training, testing and editing tools for "Bridging Global Context Interactions for High-Fidelity I
RFDesign - Protein hallucination and inpainting with RoseTTAFold
RFDesign: Protein hallucination and inpainting with RoseTTAFold Jue Wang (juewan

CHERRY is a python library for predicting the interactions between viral and prokaryotic genomes
CHERRY is a python library for predicting the interactions between viral and prokaryotic genomes. CHERRY is based on a deep learning model, which consists of a graph convolutional encoder and a link prediction decoder.

A discord.py extension for sending, receiving and handling ui interactions in discord
discord-ui A discord.py extension for using discord ui/interaction features pip package ▪ read the docs ▪ examples Introduction This is a discord.py u
A fast Protein Chain / Ligand Extractor and organizer.
Are you tired of using visualization software, or full blown suites just to separate protein chains / ligands ? Are you tired of organizing the mess o
NeoDTI: Neural integration of neighbor information from a heterogeneous network for discovering new drug-target interactions
NeoDTI NeoDTI: Neural integration of neighbor information from a heterogeneous network for discovering new drug-target interactions (Bioinformatics).
This program analyzes a DNA sequence and outputs snippets of DNA that are likely to be protein-coding genes.
This program analyzes a DNA sequence and outputs snippets of DNA that are likely to be protein-coding genes.

Boilerplate template for the discord-py-interactions library
discord-py-interactions_boilerplate Boilerplate template for the discord-py-interactions library Currently, this boilerplate supports discord-py-inter

Infrastructure template and Jupyter notebooks for running RoseTTAFold on AWS Batch.
AWS RoseTTAFold Infrastructure template and Jupyter notebooks for running RoseTTAFold on AWS Batch. Overview Proteins are large biomolecules that play
Neon: an add-on for making it easier to handle component interactions
Neon Neon is an add-on for Lightbulb making it easier to handle component interactions. Installation pip install git+https://github.com/neonjonn/light
Neon: an add-on for Lightbulb making it easier to handle component interactions
Neon Neon is an add-on for Lightbulb making it easier to handle component interactions. Installation pip install git+https://github.com/neonjonn/light
Uni-Fold: Training your own deep protein-folding models.
Uni-Fold: Training your own deep protein-folding models. This package provides and implementation of a trainable, Transformer-based deep protein foldi
Uni-Fold: Training your own deep protein-folding models
Uni-Fold: Training your own deep protein-folding models. This package provides an implementation of a trainable, Transformer-based deep protein foldin

Official PyTorch implementation for paper "Efficient Two-Stage Detection of Human–Object Interactions with a Novel Unary–Pairwise Transformer"
UPT: Unary–Pairwise Transformers This repository contains the official PyTorch implementation for the paper Frederic Z. Zhang, Dylan Campbell and Step

Decoding the Protein-ligand Interactions Using Parallel Graph Neural Networks
Decoding the Protein-ligand Interactions Using Parallel Graph Neural Networks Requirements python 0.10+ rdkit 2020.03.3.0 biopython 1.78 openbabel 2.4
A discord http interactions framework built on top of Sanic
snowfin An async discord http interactions framework built on top of Sanic Installing for now just install the package through pip via github # Unix b

A Protein-RNA Interface Predictor Based on Semantics of Sequences
PRIP PRIP:A Protein-RNA Interface Predictor Based on Semantics of Sequences installation gensim==3.8.3 matplotlib==3.1.3 xgboost==1.3.3 prettytable==2
A package to predict protein inter-residue geometries from sequence data
trRosetta This package is a part of trRosetta protein structure prediction protocol developed in: Improved protein structure prediction using predicte

Bio-Computing Platform Featuring Large-Scale Representation Learning and Multi-Task Deep Learning “螺旋桨”生物计算工具集
English | 简体中文 Latest News 2021.10.25 Paper "Docking-based Virtual Screening with Multi-Task Learning" is accepted by BIBM 2021. 2021.07.29 PaddleHeli

A small and fun Discord Bot that is written in Python and discord-interactions (with discord.py)
Articuno (discord-interactions) A small and fun Discord Bot that is written in Python and discord-interactions (with discord.py) Get started If you wa

An API wrapper for Discord written in Python.
disnake A modern, easy to use, feature-rich, and async ready API wrapper for Discord written in Python. About disnake All the contributors and develop
Create large-scale ML-driven multiscale simulation ensembles to study the interactions
MuMMI RAS v0.1 Released: Nov 16, 2021 MuMMI RAS is the application component of the MuMMI framework developed to create large-scale ML-driven multisca

Readings for "A Unified View of Relational Deep Learning for Polypharmacy Side Effect, Combination Therapy, and Drug-Drug Interaction Prediction."
Polypharmacy - DDI - Synergy Survey The Survey Paper This repository accompanies our survey paper A Unified View of Relational Deep Learning for Polyp

This package is a python library with tools for the Molecular Simulation - Software Gromos.
This package is a python library with tools for the Molecular Simulation - Software Gromos. It allows you to easily set up, manage and analyze simulations in python.
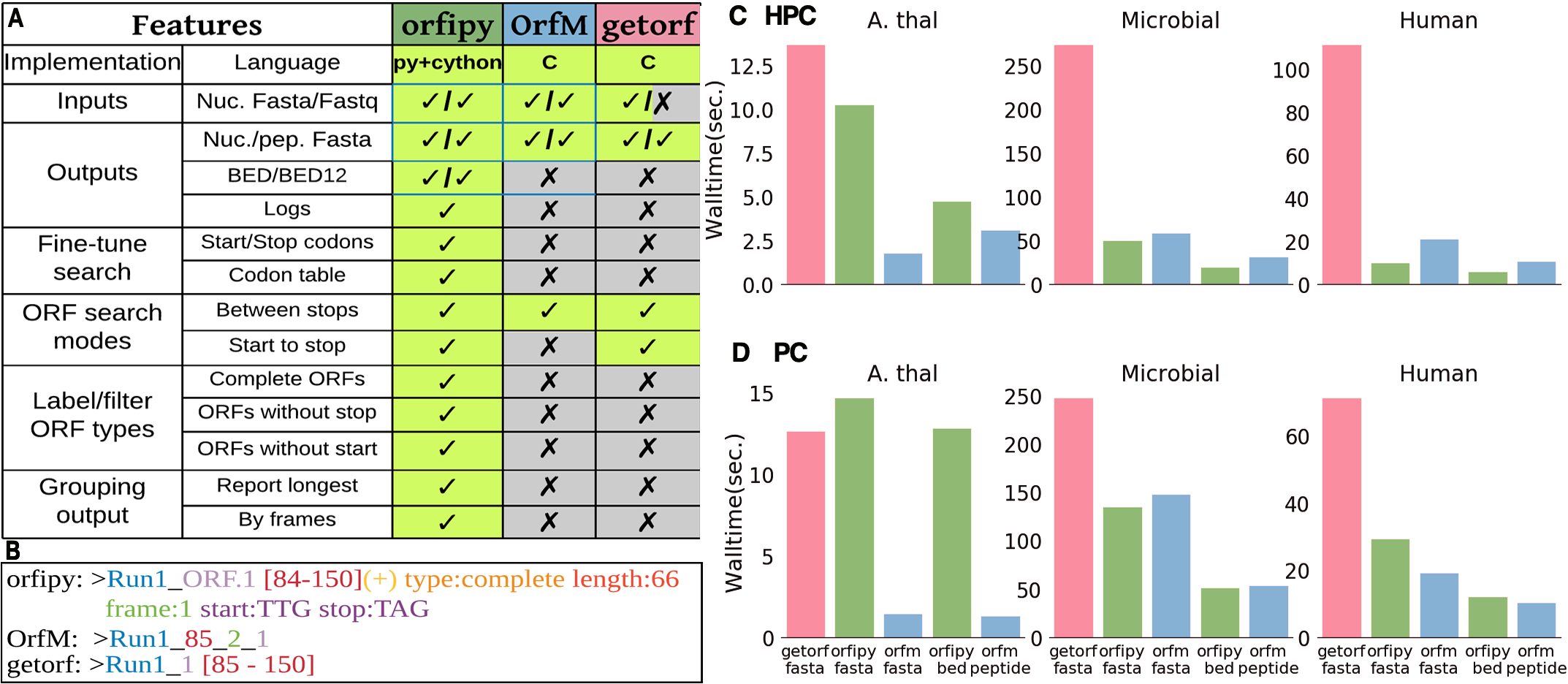
orfipy is a tool written in python/cython to extract ORFs in an extremely and fast and flexible manner
Introduction orfipy is a tool written in python/cython to extract ORFs in an extremely and fast and flexible manner. Other popular ORF searching tools

cLoops2: full stack analysis tool for chromatin interactions
cLoops2: full stack analysis tool for chromatin interactions Introduction cLoops2 is an extension of our previous work, cLoops. From loop-calling base

Bioinformatics tool for exploring RNA-Protein interactions
Explore RNA-Protein interactions. RNPFind is a bioinformatics tool. It takes an RNA transcript as input and gives a list of RNA binding protein (RBP)

Readings for "A Unified View of Relational Deep Learning for Polypharmacy Side Effect, Combination Therapy, and Drug-Drug Interaction Prediction."
Polypharmacy - DDI - Synergy Survey The Survey Paper This repository accompanies our survey paper A Unified View of Relational Deep Learning for Polyp

Code for ICMI2020 and ICMI2021 papers: "Studying Person-Specific Pointing and Gaze Behavior for Multimodal Referencing of Outside Objects from a Moving Vehicle" and "ML-PersRef: A Machine Learning-based Personalized Multimodal Fusion Approach for Referencing Outside Objects From a Moving Vehicle"
ML-PersRef This repository has python code (in jupyter notebooks) for both of the following papers: ML-PersRef: A Machine Learning-based Personalized

A simple API wrapper for Discord interactions.
Your ultimate Discord interactions library for discord.py. About | Installation | Examples | Discord | PyPI About What is discord-py-interactions? dis
Deep generative models of 3D grids for structure-based drug discovery
What is liGAN? liGAN is a research codebase for training and evaluating deep generative models for de novo drug design based on 3D atomic density grid
Predicting lncRNA–protein interactions based on graph autoencoders and collaborative training
Predicting lncRNA–protein interactions based on graph autoencoders and collaborative training Code for our paper "Predicting lncRNA–protein interactio
Unit testing AWS interactions with pytest and moto. These examples demonstrate how to structure, setup, teardown, mock, and conduct unit testing. The source code is only intended to demonstrate unit testing.
Unit Testing Interactions with Amazon Web Services (AWS) Unit testing AWS interactions with pytest and moto. These examples demonstrate how to structu
peptides.py is a pure-Python package to compute common descriptors for protein sequences
peptides.py Physicochemical properties and indices for amino-acid sequences. 🗺️ Overview peptides.py is a pure-Python package to compute common descr
Forecasting Nonverbal Social Signals during Dyadic Interactions with Generative Adversarial Neural Networks
ForecastingNonverbalSignals This is the implementation for the paper Forecasting Nonverbal Social Signals during Dyadic Interactions with Generative A

Detecting Beneficial Feature Interactions for Recommender Systems, AAAI 2021
Detecting Beneficial Feature Interactions for Recommender Systems (L0-SIGN) This is our implementation for the paper: Su, Y., Zhang, R., Erfani, S., &
Ape is a framework for Web3 Python applications and smart contracts, with advanced functionality for testing, deployment, and on-chain interactions.
Ape Framework Ape is a framework for Web3 Python applications and smart contracts, with advanced functionality for testing, deployment, and on-chain i
A graph neural network (GNN) model to predict protein-protein interactions (PPI) with no sample features
A graph neural network (GNN) model to predict protein-protein interactions (PPI) with no sample features
A geometric deep learning pipeline for predicting protein interface contacts.
A geometric deep learning pipeline for predicting protein interface contacts.
Replication attempt for the Protein Folding Model
RGN2-Replica (WIP) To eventually become an unofficial working Pytorch implementation of RGN2, an state of the art model for MSA-less Protein Folding f
This script checks for any possible SSRF dns/http interactions in xmlrpc.php pingback feature
rpckiller This script checks for any possible SSRF dns/http interactions in xmlrpc.php pingback feature and with that you can further try to escalate
Discord Panel is an AIO panel for Discord that aims to have all the needed tools related to user token interactions, as in nuking and also everything you could possibly need for raids
Discord Panel Discord Panel is an AIO panel for Discord that aims to have all the needed tools related to user token interactions, as in nuking and al

This repository contains various models targetting multimodal representation learning, multimodal fusion for downstream tasks such as multimodal sentiment analysis.
Multimodal Deep Learning 🎆 🎆 🎆 Announcing the multimodal deep learning repository that contains implementation of various deep learning-based model

Graph-based community clustering approach to extract protein domains from a predicted aligned error matrix
Using a predicted aligned error matrix corresponding to an AlphaFold2 model , returns a series of lists of residue indices, where each list corresponds to a set of residues clustering together into a pseudo-rigid domain.

Official code for "Focal Self-attention for Local-Global Interactions in Vision Transformers"
Focal Transformer This is the official implementation of our Focal Transformer -- "Focal Self-attention for Local-Global Interactions in Vision Transf

Implementation of the Triangle Multiplicative module, used in Alphafold2 as an efficient way to mix rows or columns of a 2d feature map, as a standalone package for Pytorch
Triangle Multiplicative Module - Pytorch Implementation of the Triangle Multiplicative module, used in Alphafold2 as an efficient way to mix rows or c

Implementation of Invariant Point Attention, used for coordinate refinement in the structure module of Alphafold2, as a standalone Pytorch module
Invariant Point Attention - Pytorch Implementation of Invariant Point Attention as a standalone module, which was used in the structure module of Alph

It is a temporary project to study discord interactions. You can set permissions conveniently when you invite a particular disk code bot.
Permission Bot 디스코드 내에 있는 message-components 를 연구하기 위하여 제작된 봇입니다. Setup /config/config_example.ini 파일을 /config/config.ini으로 변환합니다. config 파일의 기본 양식은 아

Vvim - Keyboardless Vim interactions
This is done via a hardware glove that the user wears. The glove detects the finger's positions and translates them into key presses. It's currently a work in progress.
Implementation and replication of ProGen, Language Modeling for Protein Generation, in Jax
ProGen - (wip) Implementation and replication of ProGen, Language Modeling for Protein Generation, in Pytorch and Jax (the weights will be made easily
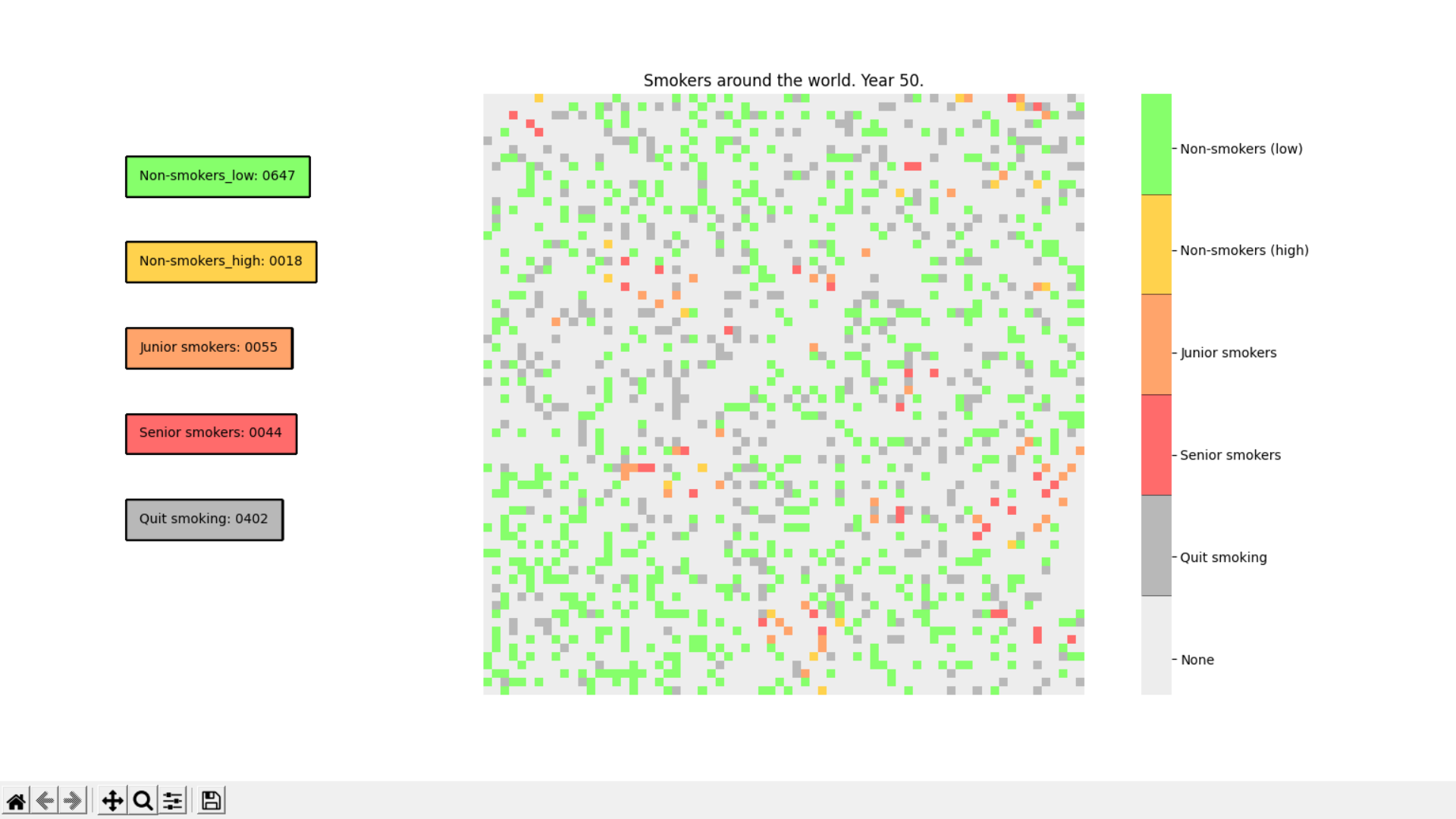
Smoking Simulation is an app to simulate the spreading of smokers and non-smokers, their interactions and population during certain amount of time.
Smoking Simulation is an app to simulate the spreading of smokers and non-smokers, their interactions and population during certain
7th place solution of Human Protein Atlas - Single Cell Classification on Kaggle
kaggle-hpa-2021-7th-place-solution Code for 7th place solution of Human Protein Atlas - Single Cell Classification on Kaggle. A description of the met

A denoising diffusion probabilistic model (DDPM) tailored for conditional generation of protein distograms
Denoising Diffusion Probabilistic Model for Proteins Implementation of Denoising Diffusion Probabilistic Model in Pytorch. It is a new approach to gen

Semi-Supervised 3D Hand-Object Poses Estimation with Interactions in Time
Semi Hand-Object Semi-Supervised 3D Hand-Object Poses Estimation with Interactions in Time (CVPR 2021).

Unofficial TensorFlow implementation of Protein Interface Prediction using Graph Convolutional Networks.
[TensorFlow] Protein Interface Prediction using Graph Convolutional Networks Unofficial TensorFlow implementation of Protein Interface Prediction usin
Protein Language Model
ProteinLM We pretrain protein language model based on Megatron-LM framework, and then evaluate the pretrained model results on TAPE (Tasks Assessing P
Implementation of ProteinBERT in Pytorch
ProteinBERT - Pytorch (wip) Implementation of ProteinBERT in Pytorch. Original Repository Install $ pip install protein-bert-pytorch Usage import torc
Implementation of ProteinBERT in Pytorch
ProteinBERT - Pytorch (wip) Implementation of ProteinBERT in Pytorch. Original Repository Install $ pip install protein-bert-pytorch Usage import torc
ProteinBERT is a universal protein language model pretrained on ~106M proteins from the UniRef90 dataset.
ProteinBERT is a universal protein language model pretrained on ~106M proteins from the UniRef90 dataset. Through its Python API, the pretrained model can be fine-tuned on any protein-related task in a matter of minutes. Based on our experiments with a wide range of benchmarks, ProteinBERT usually achieves state-of-the-art performance. ProteinBERT is built on TenforFlow/Keras.

Deep functional residue identification
DeepFRI Deep functional residue identification Citing @article {Gligorijevic2019, author = {Gligorijevic, Vladimir and Renfrew, P. Douglas and Koscio

SkipGNN: Predicting Molecular Interactions with Skip-Graph Networks (Scientific Reports)
SkipGNN: Predicting Molecular Interactions with Skip-Graph Networks Molecular interaction networks are powerful resources for the discovery. While dee

Generative Models for Graph-Based Protein Design
Graph-Based Protein Design This repo contains code for Generative Models for Graph-Based Protein Design by John Ingraham, Vikas Garg, Regina Barzilay
Structural basis for solubility in protein expression systems
Structural basis for solubility in protein expression systems Large-scale protein production for biotechnology and biopharmaceutical applications rely
git《Self-Attention Attribution: Interpreting Information Interactions Inside Transformer》(AAAI 2021) GitHub:
Self-Attention Attribution This repository contains the implementation for AAAI-2021 paper Self-Attention Attribution: Interpreting Information Intera
[CIKM 2019] Code and dataset for "Fi-GNN: Modeling Feature Interactions via Graph Neural Networks for CTR Prediction"
FiGNN for CTR prediction The code and data for our paper in CIKM2019: Fi-GNN: Modeling Feature Interactions via Graph Neural Networks for CTR Predicti
Learning Intents behind Interactions with Knowledge Graph for Recommendation, WWW2021
Learning Intents behind Interactions with Knowledge Graph for Recommendation This is our PyTorch implementation for the paper: Xiang Wang, Tinglin Hua
Differentiable molecular simulation of proteins with a coarse-grained potential
Differentiable molecular simulation of proteins with a coarse-grained potential This repository contains the learned potential, simulation scripts and
This package allows interactions with the BuyCoins API.
The BuyCoins Python library allows interactions with the BuyCoins API from applications written in Python.
📷 Instagram Bot - Tool for automated Instagram interactions
InstaPy Tooling that automates your social media interactions to “farm” Likes, Comments, and Followers on Instagram Implemented in Python using the Se
Typed interactions with the GitHub API v3
PyGitHub PyGitHub is a Python library to access the GitHub API v3 and Github Enterprise API v3. This library enables you to manage GitHub resources su
Automatically mock your HTTP interactions to simplify and speed up testing
VCR.py 📼 This is a Python version of Ruby's VCR library. Source code https://github.com/kevin1024/vcrpy Documentation https://vcrpy.readthedocs.io/ R

Implementation of trRosetta and trDesign for Pytorch, made into a convenient package
trRosetta - Pytorch (wip) Implementation of trRosetta and trDesign for Pytorch, made into a convenient package

Implementation of Geometric Vector Perceptron, a simple circuit for 3d rotation equivariance for learning over large biomolecules, in Pytorch. Idea proposed and accepted at ICLR 2021
Geometric Vector Perceptron Implementation of Geometric Vector Perceptron, a simple circuit with 3d rotation equivariance for learning over large biom
Useful tools for building interactions in Python
discord-interactions-python Types and helper functions for Discord Interactions webhooks. Installation Available via pypi: pip install discord-interac
Automatically mock your HTTP interactions to simplify and speed up testing
VCR.py 📼 This is a Python version of Ruby's VCR library. Source code https://github.com/kevin1024/vcrpy Documentation https://vcrpy.readthedocs.io/ R
Automatically mock your HTTP interactions to simplify and speed up testing
VCR.py 📼 This is a Python version of Ruby's VCR library. Source code https://github.com/kevin1024/vcrpy Documentation https://vcrpy.readthedocs.io/ R