106 Repositories
Python protein-pretraining Libraries
![[ACL 2022] LinkBERT: A Knowledgeable Language Model 😎 Pretrained with Document Links](https://github.com/michiyasunaga/LinkBERT/raw/main/figs/overview.png)
[ACL 2022] LinkBERT: A Knowledgeable Language Model 😎 Pretrained with Document Links
LinkBERT: A Knowledgeable Language Model Pretrained with Document Links This repo provides the model, code & data of our paper: LinkBERT: Pretraining

Jupyter Dock is a set of Jupyter Notebooks for performing molecular docking protocols interactively, as well as visualizing, converting file formats and analyzing the results.
Molecular Docking integrated in Jupyter Notebooks Description | Citation | Installation | Examples | Limitations | License Table of content Descriptio

BigDetection: A Large-scale Benchmark for Improved Object Detector Pre-training
BigDetection: A Large-scale Benchmark for Improved Object Detector Pre-training By Likun Cai, Zhi Zhang, Yi Zhu, Li Zhang, Mu Li, Xiangyang Xue. This

RITA is a family of autoregressive protein models, developed by LightOn in collaboration with the OATML group at Oxford and the Debora Marks Lab at Harvard.
RITA: a Study on Scaling Up Generative Protein Sequence Models RITA is a family of autoregressive protein models, developed by a collaboration of Ligh

code for TCL: Vision-Language Pre-Training with Triple Contrastive Learning, CVPR 2022
Vision-Language Pre-Training with Triple Contrastive Learning, CVPR 2022 News (03/16/2022) upload retrieval checkpoints finetuned on COCO and Flickr T

Official repository of OFA. Paper: Unifying Architectures, Tasks, and Modalities Through a Simple Sequence-to-Sequence Learning Framework
Paper | Blog OFA is a unified multimodal pretrained model that unifies modalities (i.e., cross-modality, vision, language) and tasks (e.g., image gene
![[ICLR 2022] Pretraining Text Encoders with Adversarial Mixture of Training Signal Generators](https://github.com/microsoft/AMOS/raw/main/AMOS.png)
[ICLR 2022] Pretraining Text Encoders with Adversarial Mixture of Training Signal Generators
AMOS This repository contains the scripts for fine-tuning AMOS pretrained models on GLUE and SQuAD 2.0 benchmarks. Paper: Pretraining Text Encoders wi

Commonality in Natural Images Rescues GANs: Pretraining GANs with Generic and Privacy-free Synthetic Data - Official PyTorch Implementation (CVPR 2022)
Commonality in Natural Images Rescues GANs: Pretraining GANs with Generic and Privacy-free Synthetic Data (CVPR 2022) Potentials of primitive shapes f

Implementation of a protein autoregressive language model, but with autoregressive infilling objective (editing subsequences capability)
Protein GLM (wip) Implementation of a protein autoregressive language model, but with autoregressive infilling objective (editing subsequences capabil

Implementation of the GVP-Transformer, which was used in the paper "Learning inverse folding from millions of predicted structures" for de novo protein design alongside Alphafold2
GVP Transformer (wip) Implementation of the GVP-Transformer, which was used in the paper Learning inverse folding from millions of predicted structure

Official implementation of "Generating 3D Molecules for Target Protein Binding"
Generating 3D Molecules for Target Protein Binding This is the official implementation of the GraphBP method proposed in the following paper. Meng Liu

Addon and nodes for working with structural biology and molecular data in Blender.
Molecular Nodes 🧬 🔬 💻 Buy Me a Coffee to Keep Development Going! Join a Community of Blender SciVis People! What is Molecular Nodes? Molecular Node

ProtFeat is protein feature extraction tool that utilizes POSSUM and iFeature.
Description: ProtFeat is designed to extract the protein features by employing POSSUM and iFeature python-based tools. ProtFeat includes a total of 39

Unifying Architectures, Tasks, and Modalities Through a Simple Sequence-to-Sequence Learning Framework
Official repository of OFA. Paper: Unifying Architectures, Tasks, and Modalities Through a Simple Sequence-to-Sequence Learning Framework

EquiBind: Geometric Deep Learning for Drug Binding Structure Prediction
EquiBind: geometric deep learning for fast predictions of the 3D structure in which a small molecule binds to a protein

Benchmarking Pipeline for Prediction of Protein-Protein Interactions
B4PPI Benchmarking Pipeline for the Prediction of Protein-Protein Interactions How this benchmarking pipeline has been built, and how to use it, is de
A small tool to test and visualize protein embeddings and amino acid proportions.
polyprotein_stats A small tool to test and visualize protein embeddings and amino acid proportions. Currently deployed on streamlit.io. Given a set of

Source code for EquiDock: Independent SE(3)-Equivariant Models for End-to-End Rigid Protein Docking (ICLR 2022)
Source code for EquiDock: Independent SE(3)-Equivariant Models for End-to-End Rigid Protein Docking (ICLR 2022) Please cite "Independent SE(3)-Equivar

RuCLIP tiny (Russian Contrastive Language–Image Pretraining) is a neural network trained to work with different pairs (images, texts).
RuCLIPtiny Zero-shot image classification model for Russian language RuCLIP tiny (Russian Contrastive Language–Image Pretraining) is a neural network
Codes and models for the paper "Learning Unknown from Correlations: Graph Neural Network for Inter-novel-protein Interaction Prediction".
GNN_PPI Codes and models for the paper "Learning Unknown from Correlations: Graph Neural Network for Inter-novel-protein Interaction Prediction". Lear

RuCLIP-SB (Russian Contrastive Language–Image Pretraining SWIN-BERT) is a multimodal model for obtaining images and text similarities and rearranging captions and pictures. Unlike other versions of the model we use BERT for text encoder and SWIN transformer for image encoder.
ruCLIP-SB RuCLIP-SB (Russian Contrastive Language–Image Pretraining SWIN-BERT) is a multimodal model for obtaining images and text similarities and re

OntoProtein: Protein Pretraining With Ontology Embedding
OntoProtein This is the implement of the paper "OntoProtein: Protein Pretraining With Ontology Embedding". OntoProtein is an effective method that mak
Improved Fitness Optimization Landscapes for Sequence Design
ReLSO Improved Fitness Optimization Landscapes for Sequence Design Description Citation How to run Training models Original data source Description In
PROTEIN EXPRESSION ANALYSIS FOR DOWN SYNDROME
PROTEIN-EXPRESSION-ANALYSIS-FOR-DOWN-SYNDROME Down syndrome (DS) is a chromosomal disorder where organisms have an extra chromosome 21, sometimes know
RFDesign - Protein hallucination and inpainting with RoseTTAFold
RFDesign: Protein hallucination and inpainting with RoseTTAFold Jue Wang (juewan
Pretraining on Dynamic Graph Neural Networks
Pretraining on Dynamic Graph Neural Networks Our article is PT-DGNN and the code is modified based on GPT-GNN Requirements python 3.6 Ubuntu 18.04.5 L
A fast Protein Chain / Ligand Extractor and organizer.
Are you tired of using visualization software, or full blown suites just to separate protein chains / ligands ? Are you tired of organizing the mess o

Code to generate datasets used in "How Useful is Self-Supervised Pretraining for Visual Tasks?"
Synthetic dataset rendering Framework for producing the synthetic datasets used in: How Useful is Self-Supervised Pretraining for Visual Tasks? Alejan
This program analyzes a DNA sequence and outputs snippets of DNA that are likely to be protein-coding genes.
This program analyzes a DNA sequence and outputs snippets of DNA that are likely to be protein-coding genes.
magiCARP: Contrastive Authoring+Reviewing Pretraining
magiCARP: Contrastive Authoring+Reviewing Pretraining Welcome to the magiCARP API, the test bed used by EleutherAI for performing text/text bi-encoder
This repository contains the code, models and datasets discussed in our paper "Few-Shot Question Answering by Pretraining Span Selection"
Splinter This repository contains the code, models and datasets discussed in our paper "Few-Shot Question Answering by Pretraining Span Selection", to
Large-scale pretraining for dialogue
A State-of-the-Art Large-scale Pretrained Response Generation Model (DialoGPT) This repository contains the source code and trained model for a large-

Infrastructure template and Jupyter notebooks for running RoseTTAFold on AWS Batch.
AWS RoseTTAFold Infrastructure template and Jupyter notebooks for running RoseTTAFold on AWS Batch. Overview Proteins are large biomolecules that play

Easy Language Model Pretraining leveraging Huggingface's Transformers and Datasets
Easy Language Model Pretraining leveraging Huggingface's Transformers and Datasets What is LASSL • How to Use What is LASSL LASSL은 LAnguage Semi-Super
Uni-Fold: Training your own deep protein-folding models.
Uni-Fold: Training your own deep protein-folding models. This package provides and implementation of a trainable, Transformer-based deep protein foldi
![[NeurIPS 2021] COCO-LM: Correcting and Contrasting Text Sequences for Language Model Pretraining](https://github.com/microsoft/COCO-LM/raw/main/coco-lm.png)
[NeurIPS 2021] COCO-LM: Correcting and Contrasting Text Sequences for Language Model Pretraining
COCO-LM This repository contains the scripts for fine-tuning COCO-LM pretrained models on GLUE and SQuAD 2.0 benchmarks. Paper: COCO-LM: Correcting an
Uni-Fold: Training your own deep protein-folding models
Uni-Fold: Training your own deep protein-folding models. This package provides an implementation of a trainable, Transformer-based deep protein foldin

Research Code for NeurIPS 2020 Spotlight paper "Large-Scale Adversarial Training for Vision-and-Language Representation Learning": UNITER adversarial training part
VILLA: Vision-and-Language Adversarial Training This is the official repository of VILLA (NeurIPS 2020 Spotlight). This repository currently supports

Contrastive Language-Image Pretraining
CLIP [Blog] [Paper] [Model Card] [Colab] CLIP (Contrastive Language-Image Pre-Training) is a neural network trained on a variety of (image, text) pair
Autoencoders pretraining using clustering
Autoencoders pretraining using clustering
The repository for the paper "When Do You Need Billions of Words of Pretraining Data?"
pretraining-learning-curves This is the repository for the paper When Do You Need Billions of Words of Pretraining Data? Edge Probing We use jiant1 fo
Code associated with the Don't Stop Pretraining ACL 2020 paper
dont-stop-pretraining Code associated with the Don't Stop Pretraining ACL 2020 paper Citation @inproceedings{dontstoppretraining2020, author = {Suchi

Decoding the Protein-ligand Interactions Using Parallel Graph Neural Networks
Decoding the Protein-ligand Interactions Using Parallel Graph Neural Networks Requirements python 0.10+ rdkit 2020.03.3.0 biopython 1.78 openbabel 2.4
Code accompanying paper: Meta-Learning to Improve Pre-Training
Meta-Learning to Improve Pre-Training This folder contains code to run experiments in the paper Meta-Learning to Improve Pre-Training, NeurIPS 2021. P

A Protein-RNA Interface Predictor Based on Semantics of Sequences
PRIP PRIP:A Protein-RNA Interface Predictor Based on Semantics of Sequences installation gensim==3.8.3 matplotlib==3.1.3 xgboost==1.3.3 prettytable==2
fastai ulmfit - Pretraining the Language Model, Fine-Tuning and training a Classifier
fast.ai ULMFiT with SentencePiece from pretraining to deployment Motivation: Why even bother with a non-BERT / Transformer language model? Short answe
A package to predict protein inter-residue geometries from sequence data
trRosetta This package is a part of trRosetta protein structure prediction protocol developed in: Improved protein structure prediction using predicte
XLNet: Generalized Autoregressive Pretraining for Language Understanding
Introduction XLNet is a new unsupervised language representation learning method based on a novel generalized permutation language modeling objective.

Bio-Computing Platform Featuring Large-Scale Representation Learning and Multi-Task Deep Learning “螺旋桨”生物计算工具集
English | 简体中文 Latest News 2021.10.25 Paper "Docking-based Virtual Screening with Multi-Task Learning" is accepted by BIBM 2021. 2021.07.29 PaddleHeli
Code for paper "Context-self contrastive pretraining for crop type semantic segmentation"
Code for paper "Context-self contrastive pretraining for crop type semantic segmentation" Setting up a python environment Follow the instruction in ht

Code for the TASLP paper "PSLA: Improving Audio Tagging With Pretraining, Sampling, Labeling, and Aggregation".
PSLA: Improving Audio Tagging with Pretraining, Sampling, Labeling, and Aggregation Introduction Getting Started FSD50K Recipe AudioSet Recipe Label E

TaCL: Improving BERT Pre-training with Token-aware Contrastive Learning
TaCL: Improving BERT Pre-training with Token-aware Contrastive Learning Authors: Yixuan Su, Fangyu Liu, Zaiqiao Meng, Lei Shu, Ehsan Shareghi, and Nig

TaCL: Improving BERT Pre-training with Token-aware Contrastive Learning
TaCL: Improving BERT Pre-training with Token-aware Contrastive Learning Authors: Yixuan Su, Fangyu Liu, Zaiqiao Meng, Lei Shu, Ehsan Shareghi, and Nig

NLP From Scratch Without Large-Scale Pretraining: A Simple and Efficient Framework
NLP From Scratch Without Large-Scale Pretraining This repository contains the code, pre-trained model checkpoints and curated datasets for our paper:

This package is a python library with tools for the Molecular Simulation - Software Gromos.
This package is a python library with tools for the Molecular Simulation - Software Gromos. It allows you to easily set up, manage and analyze simulations in python.
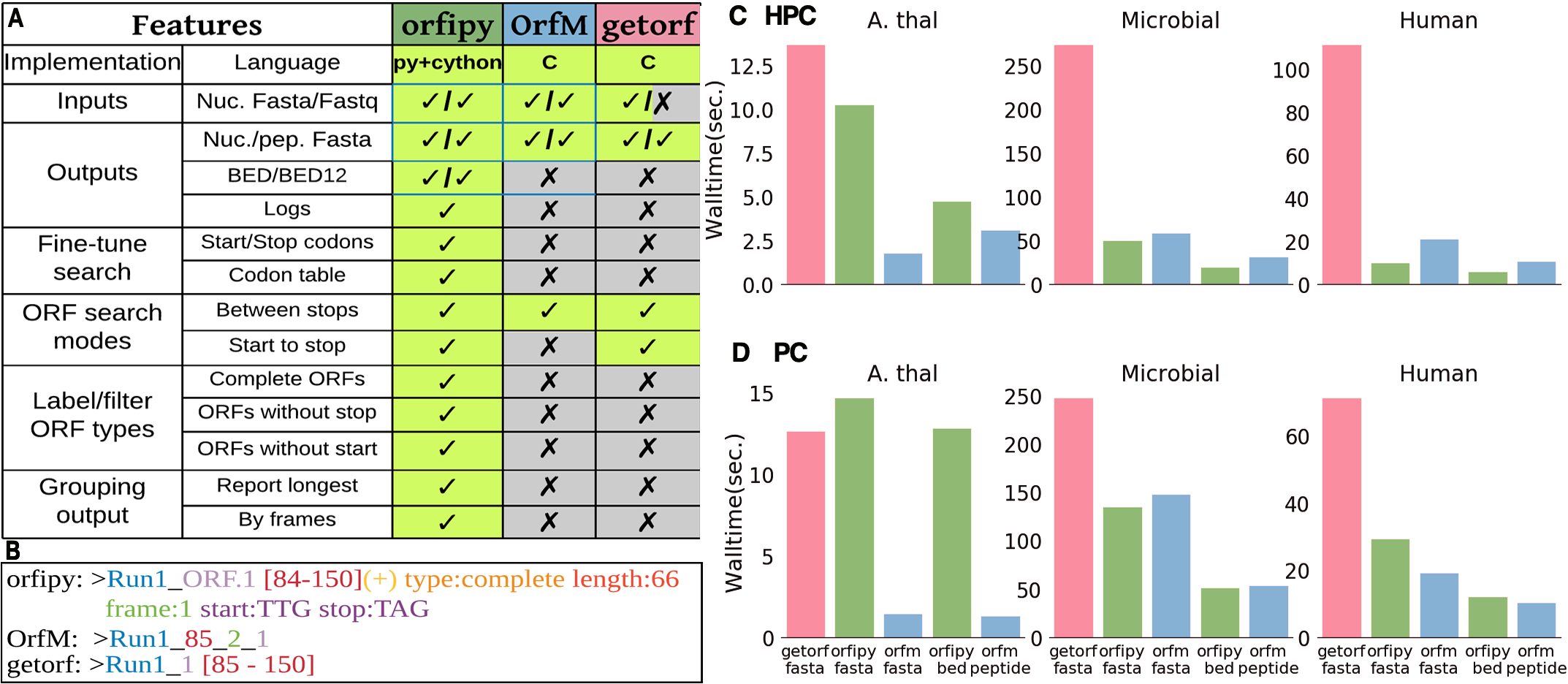
orfipy is a tool written in python/cython to extract ORFs in an extremely and fast and flexible manner
Introduction orfipy is a tool written in python/cython to extract ORFs in an extremely and fast and flexible manner. Other popular ORF searching tools

Bioinformatics tool for exploring RNA-Protein interactions
Explore RNA-Protein interactions. RNPFind is a bioinformatics tool. It takes an RNA transcript as input and gives a list of RNA binding protein (RBP)
Deep generative models of 3D grids for structure-based drug discovery
What is liGAN? liGAN is a research codebase for training and evaluating deep generative models for de novo drug design based on 3D atomic density grid
Predicting lncRNA–protein interactions based on graph autoencoders and collaborative training
Predicting lncRNA–protein interactions based on graph autoencoders and collaborative training Code for our paper "Predicting lncRNA–protein interactio
Unified Instance and Knowledge Alignment Pretraining for Aspect-based Sentiment Analysis
Unified Instance and Knowledge Alignment Pretraining for Aspect-based Sentiment Analysis Requirements python 3.7 pytorch-gpu 1.7 numpy 1.19.4 pytorch_
![[NeurIPS 2021 Spotlight] Aligning Pretraining for Detection via Object-Level Contrastive Learning](https://github.com/hologerry/SoCo/raw/main/figures/overview.png)
[NeurIPS 2021 Spotlight] Aligning Pretraining for Detection via Object-Level Contrastive Learning
SoCo [NeurIPS 2021 Spotlight] Aligning Pretraining for Detection via Object-Level Contrastive Learning By Fangyun Wei*, Yue Gao*, Zhirong Wu, Han Hu,

Code for reproducing our paper: LMSOC: An Approach for Socially Sensitive Pretraining
LMSOC: An Approach for Socially Sensitive Pretraining Code for reproducing the paper LMSOC: An Approach for Socially Sensitive Pretraining to appear a
peptides.py is a pure-Python package to compute common descriptors for protein sequences
peptides.py Physicochemical properties and indices for amino-acid sequences. 🗺️ Overview peptides.py is a pure-Python package to compute common descr
Code for Efficient Visual Pretraining with Contrastive Detection
Code for DetCon This repository contains code for the ICCV 2021 paper "Efficient Visual Pretraining with Contrastive Detection" by Olivier J. Hénaff,

Self-Supervised Speech Pre-training and Representation Learning Toolkit.
What's New Sep 2021: We host a challenge in AAAI workshop: The 2nd Self-supervised Learning for Audio and Speech Processing! See SUPERB official site
A graph neural network (GNN) model to predict protein-protein interactions (PPI) with no sample features
A graph neural network (GNN) model to predict protein-protein interactions (PPI) with no sample features
A geometric deep learning pipeline for predicting protein interface contacts.
A geometric deep learning pipeline for predicting protein interface contacts.
PyTorch original implementation of Cross-lingual Language Model Pretraining.
XLM NEW: Added XLM-R model. PyTorch original implementation of Cross-lingual Language Model Pretraining. Includes: Monolingual language model pretrain

The PASS dataset: pretrained models and how to get the data - PASS: Pictures without humAns for Self-Supervised Pretraining
The PASS dataset: pretrained models and how to get the data - PASS: Pictures without humAns for Self-Supervised Pretraining
Does Pretraining for Summarization Reuqire Knowledge Transfer?
Pretraining summarization models using a corpus of nonsense
Replication attempt for the Protein Folding Model
RGN2-Replica (WIP) To eventually become an unofficial working Pytorch implementation of RGN2, an state of the art model for MSA-less Protein Folding f
TAPEX: Table Pre-training via Learning a Neural SQL Executor
TAPEX: Table Pre-training via Learning a Neural SQL Executor The official repository which contains the code and pre-trained models for our paper TAPE
EMNLP 2021 - Frustratingly Simple Pretraining Alternatives to Masked Language Modeling
Frustratingly Simple Pretraining Alternatives to Masked Language Modeling This is the official implementation for "Frustratingly Simple Pretraining Al

Graph-based community clustering approach to extract protein domains from a predicted aligned error matrix
Using a predicted aligned error matrix corresponding to an AlphaFold2 model , returns a series of lists of residue indices, where each list corresponds to a set of residues clustering together into a pseudo-rigid domain.

X-modaler is a versatile and high-performance codebase for cross-modal analytics.
X-modaler X-modaler is a versatile and high-performance codebase for cross-modal analytics. This codebase unifies comprehensive high-quality modules i

Code for generating a single image pretraining dataset
Single Image Pretraining of Visual Representations As shown in the paper A critical analysis of self-supervision, or what we can learn from a single i

DETReg: Unsupervised Pretraining with Region Priors for Object Detection
DETReg: Unsupervised Pretraining with Region Priors for Object Detection Amir Bar, Xin Wang, Vadim Kantorov, Colorado J Reed, Roei Herzig, Gal Chechik

Implementation of the Triangle Multiplicative module, used in Alphafold2 as an efficient way to mix rows or columns of a 2d feature map, as a standalone package for Pytorch
Triangle Multiplicative Module - Pytorch Implementation of the Triangle Multiplicative module, used in Alphafold2 as an efficient way to mix rows or c
PyTorch original implementation of Cross-lingual Language Model Pretraining.
XLM NEW: Added XLM-R model. PyTorch original implementation of Cross-lingual Language Model Pretraining. Includes: Monolingual language model pretrain

Implementation of Invariant Point Attention, used for coordinate refinement in the structure module of Alphafold2, as a standalone Pytorch module
Invariant Point Attention - Pytorch Implementation of Invariant Point Attention as a standalone module, which was used in the structure module of Alph
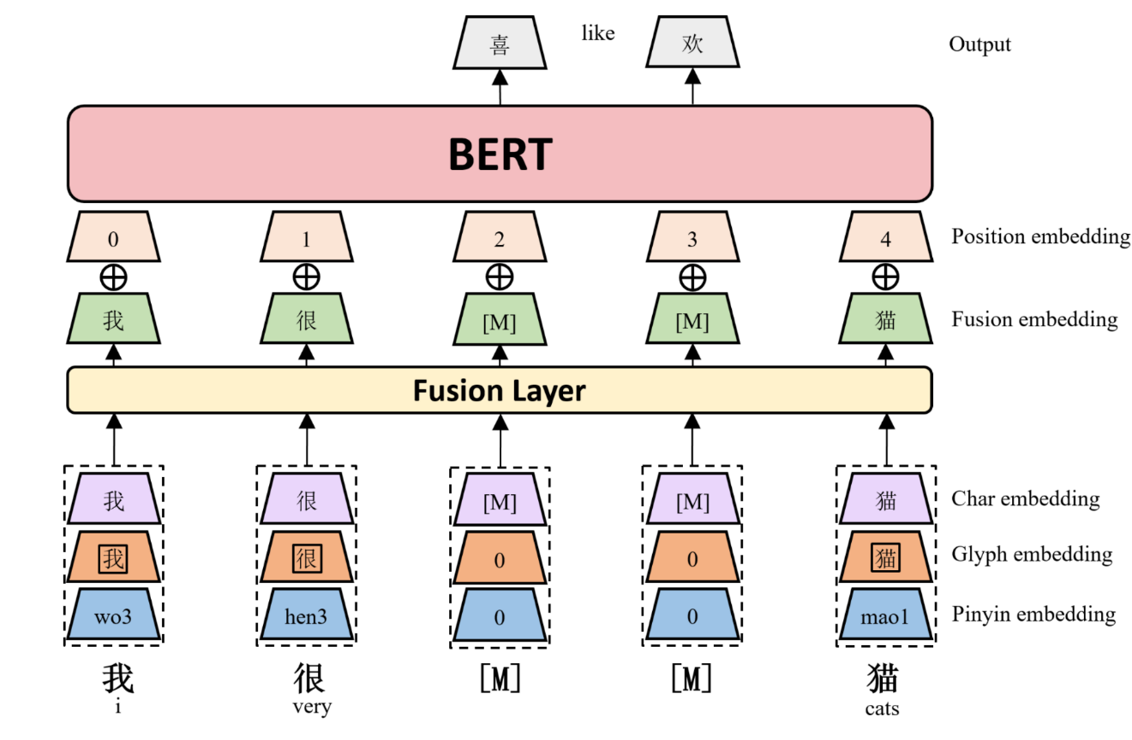
ChineseBERT: Chinese Pretraining Enhanced by Glyph and Pinyin Information
ChineseBERT: Chinese Pretraining Enhanced by Glyph and Pinyin Information This repository contains code, model, dataset for ChineseBERT at ACL2021. Ch
Implementation and replication of ProGen, Language Modeling for Protein Generation, in Jax
ProGen - (wip) Implementation and replication of ProGen, Language Modeling for Protein Generation, in Pytorch and Jax (the weights will be made easily
Pretraining Representations For Data-Efficient Reinforcement Learning
Pretraining Representations For Data-Efficient Reinforcement Learning Max Schwarzer, Nitarshan Rajkumar, Michael Noukhovitch, Ankesh Anand, Laurent Ch
7th place solution of Human Protein Atlas - Single Cell Classification on Kaggle
kaggle-hpa-2021-7th-place-solution Code for 7th place solution of Human Protein Atlas - Single Cell Classification on Kaggle. A description of the met

A denoising diffusion probabilistic model (DDPM) tailored for conditional generation of protein distograms
Denoising Diffusion Probabilistic Model for Proteins Implementation of Denoising Diffusion Probabilistic Model in Pytorch. It is a new approach to gen
![Revisiting Contrastive Methods for Unsupervised Learning of Visual Representations. [2021]](https://github.com/wvangansbeke/Revisiting-Contrastive-SSL/raw/main/images/teaser.jpg)
Revisiting Contrastive Methods for Unsupervised Learning of Visual Representations. [2021]
Revisiting Contrastive Methods for Unsupervised Learning of Visual Representations This repo contains the Pytorch implementation of our paper: Revisit

Unofficial TensorFlow implementation of Protein Interface Prediction using Graph Convolutional Networks.
[TensorFlow] Protein Interface Prediction using Graph Convolutional Networks Unofficial TensorFlow implementation of Protein Interface Prediction usin
Protein Language Model
ProteinLM We pretrain protein language model based on Megatron-LM framework, and then evaluate the pretrained model results on TAPE (Tasks Assessing P
Implementation of ProteinBERT in Pytorch
ProteinBERT - Pytorch (wip) Implementation of ProteinBERT in Pytorch. Original Repository Install $ pip install protein-bert-pytorch Usage import torc
Implementation of ProteinBERT in Pytorch
ProteinBERT - Pytorch (wip) Implementation of ProteinBERT in Pytorch. Original Repository Install $ pip install protein-bert-pytorch Usage import torc
ProteinBERT is a universal protein language model pretrained on ~106M proteins from the UniRef90 dataset.
ProteinBERT is a universal protein language model pretrained on ~106M proteins from the UniRef90 dataset. Through its Python API, the pretrained model can be fine-tuned on any protein-related task in a matter of minutes. Based on our experiments with a wide range of benchmarks, ProteinBERT usually achieves state-of-the-art performance. ProteinBERT is built on TenforFlow/Keras.

Deep functional residue identification
DeepFRI Deep functional residue identification Citing @article {Gligorijevic2019, author = {Gligorijevic, Vladimir and Renfrew, P. Douglas and Koscio

When Does Pretraining Help? Assessing Self-Supervised Learning for Law and the CaseHOLD Dataset of 53,000+ Legal Holdings
When Does Pretraining Help? Assessing Self-Supervised Learning for Law and the CaseHOLD Dataset of 53,000+ Legal Holdings This is the repository for t
![[NAACL & ACL 2021] SapBERT: Self-alignment pretraining for BERT.](https://github.com/cambridgeltl/sapbert/raw/main/sapbert_front_graphs_v6.png?raw=true)
[NAACL & ACL 2021] SapBERT: Self-alignment pretraining for BERT.
SapBERT: Self-alignment pretraining for BERT This repo holds code for the SapBERT model presented in our NAACL 2021 paper: Self-Alignment Pretraining

Generative Models for Graph-Based Protein Design
Graph-Based Protein Design This repo contains code for Generative Models for Graph-Based Protein Design by John Ingraham, Vikas Garg, Regina Barzilay
Structural basis for solubility in protein expression systems
Structural basis for solubility in protein expression systems Large-scale protein production for biotechnology and biopharmaceutical applications rely
Contrastive Language-Audio Pretraining
CLAP Contrastive Language-Audio Pretraining In due time this repo will be full of lovely things, I hope. Feel free to check out the Issues if you're i

Official Pytorch Implementation of: "ImageNet-21K Pretraining for the Masses"(2021) paper
ImageNet-21K Pretraining for the Masses Paper | Pretrained models Official PyTorch Implementation Tal Ridnik, Emanuel Ben-Baruch, Asaf Noy, Lihi Zelni

TSP: Temporally-Sensitive Pretraining of Video Encoders for Localization Tasks
TSP: Temporally-Sensitive Pretraining of Video Encoders for Localization Tasks [Paper] [Project Website] This repository holds the source code, pretra
CLASP - Contrastive Language-Aminoacid Sequence Pretraining
CLASP - Contrastive Language-Aminoacid Sequence Pretraining Repository for creating models pretrained on language and aminoacid sequences similar to C